Determining the Thermodynamic and Kinetic Association of a DNA Aptamer and Tetracycline Using Isothermal Titration Calorimetry
Summary
The present protocol describes the use of Isothermal Titration Calorimetry (ITC) to analyze the association and dissociation kinetics of the binding between a DNA aptamer and tetracycline, including sample preparation, running standards and samples, and interpreting the resulting data.
Abstract
The determination of binding affinity and behavior between an aptamer and its target is the most crucial step in selecting and using an aptamer for application. Due to the drastic differences between the aptamer and small molecules, scientists need to put much effort into characterizing their binding properties. Isothermal Titration Calorimetry (ITC) is a powerful approach for this purpose. ITC goes beyond determining disassociation constants (Kd) and can provide the enthalpy changes and binding stoichiometry of the interaction between two molecules in the solution phase. This approach conducts continuous titration using label-free molecules and records released heat over time upon the binding events produced by each titration, so the process can sensitively measure the binding between macromolecules and their small targets. Herein, the article introduces a step-by-step procedure of the ITC measurement of a selected aptamer with a small target, tetracycline. This example proves the versatility of the technique and its potential for other applications.
Introduction
Aptamers are ssDNA or RNA fragments selected through an evolution process with high binding affinity and specificity to the desired targets1,2, which can work as advanced recognition elements or chemical antibodies3,4,5. Thus, the binding affinity and specificity of aptamers to their targets play a crucial role in the selection and application of an aptamer, and Isothermal Titration Calorimetry (ITC) has been widely used for these characterization purposes. Many approaches have been used to determine the affinity of aptamers, including ITC, surface plasmon resonance (SPR), colorimetric titration, microscale thermophoresis (MST), and Bio-Layer Interferometry (BLI). Among them, ITC is one of the latest techniques to determine the thermodynamic and kinetic association of two molecules in the solution phase. This approach conducts continuous titration using label-free molecules and records released heat over time upon the binding events produced by each titration6,7. Unlike other methods, ITC can offer binding affinity, several binding sites, and thermodynamic and kinetic association (Figure 1A). From these initial parameters, the Gibbs free energy changes and entropy changes are determined using the following relationship:
ΔG = ΔH-TΔS
That means that ITC offers a complete thermodynamic profile of the molecular interaction to elucidate the binding mechanisms (Figure 1B). Determining the binding affinity for small molecules with an aptamer is difficult due to the drastically different sizes between aptamer and target. Meanwhile, ITC can provide sensitive measurement without labeling and immobilizing molecules, which provides a means of keeping the natural structure of the aptamer and target during measurement. With the mentioned attributes, ITC can be used as the standard method for the characterization of binding between an aptamer and small targets.
After selection by the Gu group, this aptamer was integrated with different platforms, including electrochemical aptamer-based biosensors, a competitive enzyme-linked aptamer assay, and a microtiter plate, which can achieve high-throughput detection of tetracycline8,9,10. However, its binding characteristics have not been elucidated well enough to choose the proper platform8; it is worth characterizing the binding of the aptamer to the tetracycline using ITC.
Protocol
NOTE: Figure 2 shows the main steps of the ITC experiment for determining the thermodynamic and kinetic association of a DNA aptamer and tetracycline.
1. Preparation of samples
NOTE: Samples for ITC need to be prepared in the same buffer for both the aptamer and ligand to avoid heat release caused by mixing different buffers from the sample cell and syringe. This is typically achieved through dialysis of all materials into the same buffer. The buffer is exchanged using a protocol adapted from the protocol of a 3 kDa molecular weight cutoff (MWC) concentrator with some modifications, as below:
- Activate the membrane of the dialysis column (3 kDa MWC) with 1x PBS, pH 7.4, purchased from the manufacturer, using the following steps: fill with 1x buffer (PBS), equilibrate for 10 min at RT, and centrifuge at 5,000 x g for 15 min.
- Remove the buffer and load 500 µL of aptamer samples into the column, centrifuge at 5,000 x g, and repeat it 4x to exchange the original buffer for 1x PBS. When the buffer goes through the membrane, all molecules with a mass less than 3 kDa will go through the membrane, and the aptamer will remain on the upper side of the membrane.
- Collect the dialyzed DNA aptamer using a pipet and transfer it to the new 1.5 mL tube(s).
- Collect the last flowthrough buffer to dissolve tetracycline. Tetracycline powder is pure and small, so dialysis is not needed. However, use the previous dialyzed buffer for DNA for the target to ensure that the buffer for the experiment in the syringe matches the buffer in the reference cell.
- Determine the aptamer concentration again using a UV-visible spectrometer. Use the last exchange buffer to adjust the concentration to 40 µM tetracycline and 2 µM aptamer.
- Fold the DNA aptamer by heating at 90 °C for 10 min, cooling at 4 °C for 10 min, and then returning to RT for 20 min.
- Degas the folded aptamer and dialyzed tetracycline using a degassing station or vacuum pump set to 600 mmHg at 25 °C for 25 min to eliminate dissolved gases.
2. Washing the instrument and running the test kit
- Clean the solvent ports to ensure the entire sample path is clear. Clean by discarding the waste solution and loading them with pure methanol, water, and buffer. Each port contains more than 250 mL to ensure enough solution for cleaning.
NOTE: The cleaning process is automatically completed by user-programmable ITC control software. - Test the cleanliness of the machine by running ITC using buffer into a buffer (i.e., 1x PBS into 1x PBS).
NOTE: A normal noise baseline is visible between the tiny buffer into buffer injection peaks. When the titration syringe and cannulas are adequately cleaned and completely dry, the baseline will be stable; an increase or decrease in the baseline reflects dirty instrumentation or bubbles inside the instrument, which need to be corrected before running actual samples. - Test the accuracy of the machine with a standard kit that includes EDTA and CaCl2 (Figure 3), using the default program and following the instructions from the manufacturer.
3. Running the sample to determine the binding between aptamer and tetracycline
- Set up the running parameters: a stirring rate of 200 rpm, running at 25 °C, 2 µM aptamer and 40 µM tetracycline, 30 injections with 2.0 µL each, a delaying time of 180 s.
- Check the required volumes using a running program calculator. With this running parameter, perform ITC measurement with 230 µL of 40 µM tetracycline in the ITC syringe and 485 µL of 2 µM aptamer in the ITC sample cell using the ITC.
- Load the dialyzed tetracycline syringe plates and the folded aptamer into the sample cell, avoiding bubbles, using a pipette.
- Start running the ITC instrument by clicking on the start button on the software.
NOTE: The ITC instrument running process is fully automated after manually filling the reference cell and titrant sample plates.
4. Analyzing data using software
- Open the data analysis software by double clicking to start analyzing the data.
- Open the path of the saved raw data to know the tendency of binding.
- Open the modeling tab and use different binding models to find the best fit for the data curve. Then, the software automatically calculates the ITC thermogram and various thermodynamic parameters, including enthalpy (ΔH), entropy (ΔS), free energy (ΔG), equilibrium binding constant (Ka), and stoichiometry.
- Collect the thermodynamic parameters determined from the data and fitting model information.
- Create a report, including pictures of the ITC thermogram and various thermodynamic parameters, as shown in Figure 4 and Table 1.
Representative Results
ITC provides an accurate disassociation constant (Kd), the binding stoichiometry, and the thermodynamic parameters of two-molecule interactions6. In this example, the aptamer selected by Kim et al.9,11 binds to tetracycline with binding affinities of Kd 1 = 13 µM, Kd 2 = 53 nM. Interestingly, this binding was determined using the equilibrium filtration method and a reported Kd of 63.3 nM, which is not much different from the favorable binding site (site 2). The fitting model and stoichiometry from ITC reflect that the aptamer binds to tetracycline through a 2:1 binding ratio with the sequential binding model (Figure 4, Table 1).
The thermodynamic parameters determined by ITC measurement for site 2 (ΔH = -1200 kcal/mol and -TΔS = 99.75 kcal/mol) indicated that enthalpy overcoming relatively significant entropic loss drives the strong binding. The enthalpy-driven binding with entropy loss relates to the RNA-conformational changes, which have been reported as binding behaviors between RNA and a small molecule. For example, Thoa et al. reported such binding behavior (ΔH = -27 kcal/mol and -TΔS = +17 kcal/mol) between an RNA aptamer and Ru(bpy)312. Besides, Horowitz et al. indicated that entropy-driven binding is associated with the intercalation of proflavine to an oligonucleotide (ΔH = -2.6 kcal/mol and -TΔS = -3.3 kcal/mol)13. Based on these comparisons, the aptamer functions with switching-structural behavior upon enthalpy-driven binding, allowing to use of the aptamer as recognition for the development of a straightforward sensor.
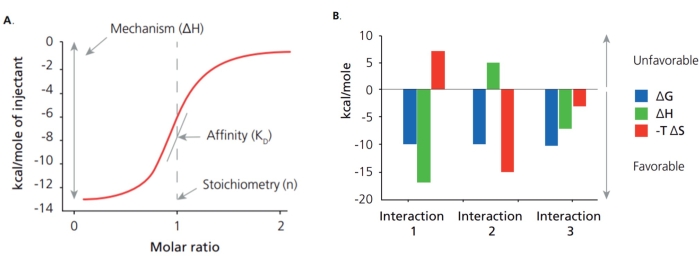
Figure 1: Binding disassociation constant (Kd) and thermodynamic profile. (A) ITC identifies the binding disassociation constant (Kd) and thermodynamic profile, including change in enthalpy (ΔH) and change in entropy (ΔS). (B) The thermodynamic profile provides the strength and mechanism of interaction. Please click here to view a larger version of this figure.

Figure 2: Main steps of the ITC experiment. The schematic shows the main steps of the ITC experiment for determining the thermodynamic and kinetic association of a DNA aptamer and tetracycline. Please click here to view a larger version of this figure.
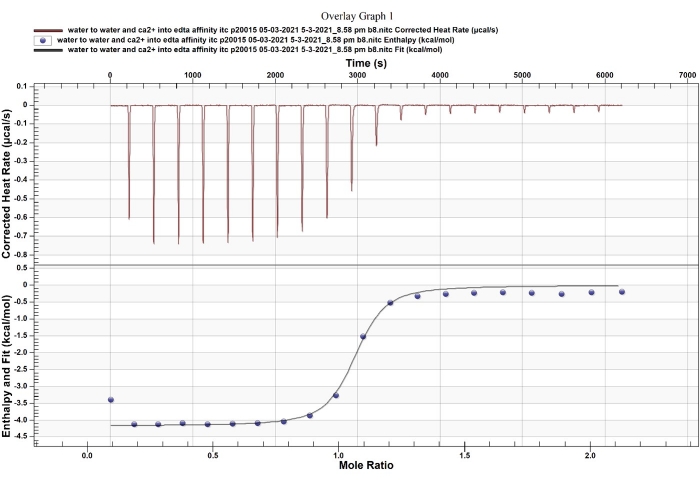
Figure 3: ITC standard test between EDTA and CaCl2. The Ca2+-EDTA chelation has been used as a standard reaction to validate ITC. Please click here to view a larger version of this figure.
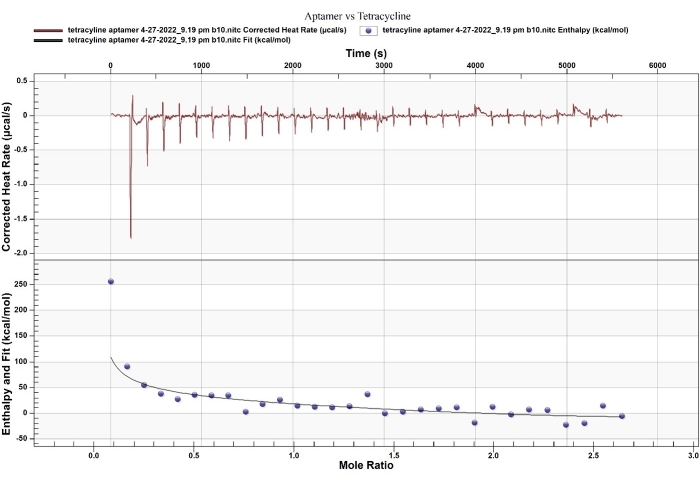
Figure 4: ITC thermogram of a DNA aptamer and tetracycline. The fitting model of thermodynamic and kinetic association reflects that the binding has two independent binding sites. Please click here to view a larger version of this figure.
| Sequential Two Site | Kd (M) | 1.359 x 10-5 |
| Kd (M) | 5.378 x 10-8 | |
| ΔH (kcal/mol) | 1223 | |
| ΔH (kcal/mol) | -1200 | |
| Ka (Mˉ¹) | 7.358 x 104 | |
| Ka (Mˉ¹) | 1.859 x 107 | |
| ΔS (cal/mol K) | 4.123 x 103 | |
| ΔS (cal/mol K) | -3.992 x 103 |
Table 1: Parameters of the binding between aptamer and tetracycline. Various thermodynamic parameters, including enthalpy (ΔH), entropy (ΔS), free energy (ΔG), equilibrium binding constant (Ka), and stoichiometry, can be determined by the binding mechanism of two molecules.
Discussion
The method presented here was modified according to instruction from TA Instruments and is sufficient to determine the binding affinity and thermodynamics of many selected aptamers and targets at our center. Crucial steps from this procedure include exchanging the buffer to have a target matching the ligand, running samples with proper parameters, and finding the appropriate binding fitting model to analyze the data. Continuous recording of heat release requires eliminating all noise heat, such as from mismatch of the buffer, dirtiness of the cell and syringe, and bubbles inside the samples. In the buffer exchange step, it is better to use the last dialysis buffer or flow-through buffer in the dialysis membrane or spin column to dissolve the ligand because it is expensive to exchange small molecules directly.
Most other binding affinity determination methods assume a 1:1 binding ratio between aptamer and ligand. However, due to the binding behavior and the drastically different size between small molecules and aptamers, the 1:1 binding model is not always accurate14,15. In this aspect, the ITC can give data on the binding stoichiometry to know the number of binding sites and give information about binding behavior7,15. That advanced function can be provided by using the correct binding model from the ITC analysis software, either the one- or two-site binding model. One saturated point can be analyzed at a different binding ratio of 1:1 (1 binding site), 1:2, or 0.5:1 (two binding sites). For the sequential model, there is no distinct saturated site but only a total number of saturated sites. If the sites are identical, the data fit with sequential saturation. There, the first binding site has more empty copies of the same kind to choose from than the second site, as evident from the decrease in released heat energy from site to site14,15,16. The binding parameters determine the binding constant K for each binding site. In this case, it shows the binding with two sequential binding sites, confirming the conformational change in the overall molecule. Although ITC offers many advanced functions to characterize the binding between the aptamer and small molecules, the optimization process having good conditions costs time7,16,17. Besides, in comparison with other instruments, ITC equipment is expensive and requires handling by a well-trained technician.
In most cases, determining the binding affinity between an aptamer and a small molecule ligand is challenging. ITC can be considered an advanced method for this purpose because ITC provides highly accurate binding affinities, as well as thermodynamic information. From this information, we can predict its behavior to use it for clinical use or detection. For example, with the selected aptamer with two binding sites, we can truncate it to keep one binding site if one site is not favorable for binding, or we can split the aptamer into two aptamers if both binding sites have the same behavior. Also, with conformation structure change behavior, we can combine the aptamer with a quenching platform to develop a sensor.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by the Research and Development Funding from Aptagen LLC.
Materials
| 5'-CGTACGGAATTCG CTAGCCCCCCGGCAGGCCACGG C TTGGGTTGGTCCCACTGCGCG TGGATCCGAGCTCCAC GTG-3' |
Integrated DNA Technologies, Inc | The sequence is adopted from Gu's research, which has not identified Kd using ITC (refer references 8 and 9) | |
| Affinity ITC Auto Low Volume (190 µL) System Complete–Gold Cells | TA Instruments | 61000.901 | Isothermal titration calorimetry system |
| CaCl2 | Avantor (VWR) | E506-100ML | Calcium chloride 1 M in aqueous solution, Biotechnology Grade, sterile |
| Centrifuge | Eppendorf | 5417R | The Eppendorf 5417R is unsurpassed in safety, reliability and ease-of-use. Very easy to maintain with a brushless motor that spins up to 16,400 RPM with maximum RCF up to 25,000 x g. |
| Complete Degassing Station (110/230V) | TA Instruments | 6326 | This degasser provides a self-contained stirring platform, vacuum chamber, vacuum port, temperature control and electronic timer for proper sample preparation. |
| EDTA | TekNova | E0375 | EDTA 500 mM, pH 7.5 |
| NanoDrop One Microvolume UV-Vis Spectrophotometer | ThermoFisher | ND-ONE-W | UV-Vis Spectrophotometer |
| Nanosep, Nanosep MF and NAB Centrifugal Devices | Pall Laboratory | OD030C34 | 3 kDa molecular weight cutoff concentrator |
| PBS pH 7.4 | IBI Scientific | IB70165 | Buffer containing Sodium phosphate, Sodium chloride, Potassium phosphate, and Potassium chloride Ultra-Pure Grade Sterile filtered using 0.2 µm filter. Autoclaved at 121 °C for greater than 20 min. |
| Posi-Click 1.7 mL Large Cap Microcentrifuge Tubes | labForce (a Thomas Scientific Brand) | 1149K01 | |
| Tetracycline, Hydrochoride | EMD Millipore Corperation | CAS64-75-5 |
References
- Ellington, A. D., Szostak, J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 346 (6287), 818-822 (1990).
- Tuerk, C., Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 249 (4968), 505-510 (1990).
- Kim, S. H., Thoa, T. T. T., Gu, M. B. Aptasensors for environmental monitoring of contaminants in water and soil. Current Opinion in Environmental Science & Health. 10, 9-21 (2019).
- Dunn, M. R., Jimenez, R. M., Chaput, J. C. Analysis of aptamer discovery and technology. Nature Reviews Chemistry. 1, 0076 (2017).
- Stoltenburg, R., Reinemann, C., Strehlitz, B. SELEX–A (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomolecular Engineering. 24 (4), 381-403 (2007).
- Wang, Y., Wang, G., Moitessier, N., Mittermaier, A. K. Enzyme kinetics by isothermal titration calorimetry: Allostery, inhibition, and dynamics. Frontiers in Molecular Biosciences. 7, 583826 (2020).
- Velazquez-Campoy, A., Freire, E. Isothermal titration calorimetry to determine association constants for high-affinity ligands. Nature Protocols. 1 (1), 186-191 (2006).
- Niazi, J. H., Lee, S. J., Gu, M. B. Single-stranded DNA aptamers specific for antibiotics tetracyclines. Bioorganic and Medicinal Chemistry. 16 (15), 7245-7253 (2008).
- Kim, Y. J., Kim, Y. S., Niazi, J. H., Gu, M. B. Electrochemical aptasensor for tetracycline detection. Bioprocess and Biosystems Engineering. 33 (1), 31-37 (2010).
- Wang, S., et al. Development of an indirect competitive assay-based aptasensor for highly sensitive detection of tetracycline residue in honey. Biosensors & Bioelectronics. 57, 192-198 (2014).
- Kim, Y. S., et al. A novel colorimetric aptasensor using gold nanoparticle for a highly sensitive and specific detection of oxytetracycline. Biosensors & Bioelectronics. 26 (4), 1644-1649 (2010).
- Thoa, T. T., Minagawa, N., Aigaki, T., Ito, Y., Uzawa, T. Regulation of photosensitisation processes by an RNA aptamer. Scientific Reports. 7, 43272 (2017).
- Horowitz, E. D., Lilavivat, S., Holladay, B. W., Germann, M. W., Hud, N. V. Solution structure and thermodynamics of 2′,5′ RNA intercalation. Journal of the American Chemical Society. 131 (16), 5831-5838 (2009).
- Sigurskjold, B. W. Exact analysis of competition ligand binding by displacement isothermal titration calorimetry. Analytical Biochemistry. 277 (2), 260-266 (2000).
- Neves, M. A. D., Slavkovic, S., Churcher, Z. R., Johnson, P. E. Salt-mediated two-site ligand binding by the cocaine-binding aptamer. Nucleic Acids Research. 45 (3), 1041-1048 (2017).
- Turnbull, W. B., Daranas, A. H. On the value of c: Can low affinity systems be studied by isothermal titration calorimetry. Journal of the American Chemical Society. 125 (48), 14859-14866 (2003).
- Van Ness, J., Van Ness, L. K., Galas, D. J. Isothermal reactions for the amplification of oligonucleotides. Proceedings of the National Academy of Sciences of the United States of America. 100 (8), 4504-4509 (2003).

