Scalable Transfection of Maize Mesophyll Protoplasts
Summary
Here, we present a high-throughput protocol to transiently transfect millions of maize protoplasts for testing large libraries of plasmids in maize mesophyll cells.
Abstract
The transfection of maize mesophyll cells often involves digesting the plant cell walls to create protoplasts and then inserting DNA via electroporation or polyethylene glycol (PEG). Previous methods were developed to produce tens of thousands of transfected protoplasts at once. Here, we describe a straightforward method to isolate and transfect millions of leaf mesophyll protoplasts in maize (Zea mays L.). This streamlined process removes certain common protoplasting steps, such as washing in W5. Additionally, steps such as centrifugation, PEG-mediated transfection, and incubation have been modified to work with a greater number of protoplasts. The ability to express large libraries of plasmid constructs enables genome-scale experiments, such as massively parallel reporter assays in maize.
Introduction
Transient gene expression is a powerful tool in plant biology. However, plant cells are difficult to transfect because they are surrounded by a cell wall. This barrier to DNA entry is not present in other commonly studied cell types such as mammalian or insect cells. Past research addressed this challenge by employing protoplasts: plant cells whose cell walls have been digested and removed. In contrast to unprocessed leaf tissue, plant protoplasts are easy to work with in suspension and amenable to transfection techniques mediated by electroporation or polyethylene glycol (PEG)1. Plant cells retain much of their functionality even after cell wall removal. Thus, protoplasts are used in a variety of plants to study gene and protein function2,3, to understand signal transduction4, and to identify possible targets for plant breeding5. Protoplasts are also a convenient vehicle for screening genome editing constructs in diverse crop species including wheat and potato6,7. In short, transient assays in protoplasts are indispensable for agricultural biotechnology.
Maize (Zea mays L.) is the world-leading cereal crop by tonnage; as such, it is a major focus of basic and applied plant science research8. However, unlike in model plants such as Nicotiana tabacum9, transient transgene expression in intact maize leaves is not routinely performed. Protoplasting has been used to obtain and transfect maize leaf mesophyll cells10,11. Previous methods have achieved transfection efficiencies up to 75% using electroporation and produced transfected protoplasts at the scale of tens of thousands10,15.
This protocol details how to protoplast millions of maize mesophyll cells and transfect them with an efficiency comparable to previous methods. The main steps are growing etiolated maize seedlings, harvesting the leaf tissue, digesting the plant cell wall, and delivering plasmid DNA into the cells using PEG. The key contribution of this protocol is the ability to scale up protoplast transfection while maintaining the cell viability required for functional assays. This enables the use of genome-scale experiments, such as massively parallel reporter assays, that can test the function of hundreds of thousands of regions throughout the maize genome. This method has recently been used to investigate large libraries of cis-regulatory DNA elements including enhancers and promoters12,13,14.
Protocol
1. Growth of the plant material
- Rinse maize kernels twice with tap water. Soak the kernels in tap water overnight at room temperature.
- The next day, fill a 72 cell seed starter tray with wetted 1:1 vermiculite:peat moss. Rinse the kernels once and plant them tip cap down, one kernel per cell.
- Grow under long-day conditions (16 h light, 8 h dark) at 25 °C for 3 days.
- Transfer to constant darkness at 25 °C, and grow for 9-11 days.
2. Plasmid isolation
- Transform commercial chemically competent E. coli with the plasmid(s) of interest according to the manufacturer's instructions.
- Grow the transformed E. coli in 50 mL of liquid LB medium with appropriate antibiotics overnight at 37 °C with shaking.
- Pellet the E. coli by centrifuging for 10 min at 3,000 x g at room temperature. Extract the plasmids using a commercially available midiprep plasmid DNA isolation kit.
- Estimate the plasmid DNA concentration using a microvolume spectrophotometer.
NOTE: Material suitable for transfection should have a concentration of ≥800 ng/µL and an A260/A280 of ≥1.8.
3. Protoplast isolation
- Prepare 20 mL of enzyme solution.
- Add 2.18 g of mannitol, 0.30 g of cellulase R-10, and 0.06 g of macerozyme R-10 to a 50 mL centrifuge tube. Invert to mix the powders. Add deionized H2O to 15 mL. Add 200 µL of 1 M MES, pH 5.7. Vortex to mix.
- Heat the solution at 55 °C for 10 min. Cool at room temperature for 10 min.
- Add 20 µL of 1 M CaCl2, 0.02 g of bovine serum albumin, and 100 µL of 1 M β-mercaptoethanol.
- Fill to 20 mL with deionized H2O. Pour into a foil-wrapped 250 mL beaker.
- Take the maize seedlings out of the dark. Cut them a few centimeters above the soil using a new razor blade. Place the cut ends in water. Keep them in the dark at room temperature while not in use.
- Select the second and third leaves of each seedling, taking care to exclude leaves with brown or damaged areas. Select 12-16 leaves. Cut ~1 cm off the tip of each leaf and discard. Then, cut a 10 cm section from each leaf.
NOTE: If more tissue is needed, use an additional 20 mL volume of enzyme solution in an additional 250 mL beaker. Too much leaf material in a given amount of enzyme solution can decrease the viability. - Stack the leaf sections on top of each other with the basal ends together. Clamp the bundle together at the basal end using a binder clip. Place the bundle of leaves on a piece of white paper.
- Grip the leaves ~1 cm from the distal end. Using a new razor blade, cut the leaves from the distal end perpendicular to the veins into thin (0.5-1 mm) slices. Avoid chopping straight down, but rather make short, forward slices through the tissue.
- Swap the razor blades whenever visible residue begins to be deposited on the paper. Visible residue indicates mashing of the tissue due to a dull blade or poor technique.
- After cutting ~2 cm of the length of the leaf bundle, promptly transfer the slices to the beaker of enzyme solution. Do not allow the material to dry out, as this will reduce the number of protoplasts obtained. Gently swirl the enzyme solution to wet the material.
- Place a foil lid over the beaker to block light. Repeat the slicing and transferring process until the entire bundle is cut close to the binder clip.
- Transfer the beaker of enzyme solution to a bell jar. Vacuum-infiltrate between −50.8 kPa and −67.7 kPa relative to ambient pressure for 3 min at room temperature.
- Transfer the beakerto a tabletop shaker. Shake at 40 RPM for 2.5 h at room temperature. Gently swirl the beaker by hand. When fully digested, the enzyme solution will appear slightly milky. If the solution is still clear, continue shaking at 40 RPM for up to one additional hour. Do not exceed 3.5 h.
- During digestion, make a mannitol + MES + MgCl2 (MMG) solution, a polyethylene glycol (PEG) solution, and an incubation solution.
- MMG: Add 5.47 g of mannitol, 200 µL of 1 M MES, pH 5.7, and 750 µL of 1 M MgCl2 to a 50 mL centrifuge tube. Fill to 50 mL with distilled H2O. Vortex to dissolve. Place on ice.
- Incubation solution: Add 5.47 g of mannitol, 200 µL of 1 M MES at pH 5.7, and 200 µL of 1 M KCl to a 50 mL centrifuge tube. Fill to 50 mL with distilled H2O. Vortex to dissolve. Place on ice.
- PEG solution: Add 0.44 g of mannitol, 1.0 g of PEG, and 400 µL of 1 M CaCl2 to a 15 mL centrifuge tube. Fill to 4 mL with distilled H2O. Vortex to mix. Incubate at 37 °C with shaking for 30 min until fully dissolved. Keep at room temperature.
- Shake the enzyme solution at 80 RPM for 10 min at room temperature.
- Place the beaker with the enzyme solution on ice. Do not remove the foil covering. Add 20 mL of ice-cold MMG to the enzyme solution. Filter through a 40 µm cell strainer into a 50 mL centrifuge tube.
NOTE: Keep all the solutions on ice for the following steps until the addition of the PEG. Keep the protoplasts covered with foil to minimize light exposure. - Split the filtrate into equal volumes of 10 mL each. Pour each volume into a round-bottom glass centrifuge tube, and spin down for 4 min at 100 x g at room temperature.
- Discard the supernatant and add 1 mL of ice-cold MMG to each tube. Resuspend the pellets by gently swirling and tapping the tubes. Combine the samples into a single round-bottom glass centrifuge tube. Add MMG to a total volume of 5 mL. Spin down for 3 min at 100 x g at room temperature.
- Wash with 5 mL of MMG, and spin down for 3 min at 100 x g at room temperature.
- Repeat the wash with 5 mL of MMG, and spin down for 3 min at 100 x g at room temperature. After spinning down the second wash, observe whether the supernatant is clear. If it remains cloudy, perform a third wash.
- Resuspend the protoplasts in 1 mL of MMG. Cover loosely with foil and keep on ice.
- Determine the protoplast yield and check the viability.
- In a 1.5 mL microcentrifuge tube, add together 4 µL of protoplasts and 72 µL of MMG.
- In a 1.5 mL microcentrifuge tube, add 1 µL of 0.5% fluorescein diacetate (FDA) stock solution to 49 µL of MMG to make a 20x working solution. Add 4 µL of FDA working solution to the tube with the diluted protoplasts. Incubate for 5 min in the dark at room temperature.
- Load 11 µL of the protoplast mixture onto a hemocytometer. Place under a brightfield light microscope. Visually confirm that the cells lack cell walls. Then, count the number of protoplasts. Use this count to estimate the total number of protoplasts obtained. Then, count the number of protoplasts that fluoresce green under UV light when stained with FDA. Successful protoplast isolations yield a viability of 70%-90%.
4. PEG-mediated transfection
- Starting with the estimated protoplast concentration from step 3.17, adjust to ~10,000 protoplasts/µL. If the concentration is low, centrifuge for 3 min at 100 x g, and resuspend in MMG to a concentration of ~10,000 protoplasts/µL.
- Mix ~1,000,000 protoplasts with 15 µg of DNA in a 1.5 mL microcentrifuge tube. Top up the mixture with MMG to 114.4 µL, and incubate on ice for 30 min.
- Resuspend the protoplasts by tapping the side of the tube. Add 105.6 µL of 25% PEG solution to the protoplasts to reach a final weight/volume of 12% PEG. Gently mix by inverting 5-10 times. Protoplasts in PEG solution are fragile; do not shake the tube. Incubate for 10 min in the dark at room temperature.
NOTE: The transfection can be scaled up in multiples of the volumes given in steps 4.2 and step 4.3. For example, 10 million protoplasts and 150 µg of DNA can be suspended in 1,444 µL of MMG and treated with 1,056 µL PEG solution in a 50 mL centrifuge tube. - Dilute with 5 volumes of incubation solution. Gently mix by inversion. Spin down for 4 min at 100 x g at room temperature.
NOTE: If scaling up, split the protoplasts into multiple round-bottom glass tubes containing no more than 10 mL each to ensure complete pelleting. Proportionately scale up the incubation solution volume used for washing. - Remove the supernatant by pipetting. Wash with 1-5 mL of incubation solution, and spin down for 3 min at 100 x g at room temperature.
- Resuspend the protoplasts in the incubation solution to a concentration of 500-1,000 cells/µL.
- Incubate the protoplasts in the dark overnight (~16 h) at room temperature.
5. Protoplast recovery and expression verification
- Spin down the protoplasts for 4 min at 100 x g at room temperature. NOTE: If scaling up, split the protoplasts into multiple round-bottom glass tubes containing no more than 10 mL each.
- Wash with 1-5 mL of incubation solution, and spin down for 3 min at 100 x g at room temperature.
- Resuspend, and combine in 1-5 mL of incubation solution.
- If the protoplasts are transfected with a fluorescent reporter gene (e.g., GFP), take an aliquot to check the transfection efficiency. Using a hemocytometer, first count the total number of protoplasts using brightfield light microscopy. Then, count the fraction of fluorescent protoplasts under UV light.
- Centrifuge the protoplasts for 3 min at 100 x g at room temperature.
NOTE: The protoplasts are now ready for downstream applications such as RNA extraction with phenol and guanidine isothiocyanate.
Representative Results
The tissue best suited for protoplast isolation is the second and third leaves of 10-11 day old seedlings (Figure 1). In this work, we obtained approximately 10 million protoplasts from 16 leaf sections, each of which was 10 cm long. The number of protoplasts isolated is dependent on the mass of the leaf material and the width of the leaf slices that are digested in the enzyme solution.
Under a brightfield light microscope, undamaged protoplasts appear roughly spherical, with plastids speckling the surface (Figure 2A). Before proceeding to transfection, the viability can be checked by staining a small aliquot of protoplasts with fluorescein diacetate (FDA). FDA-stained protoplasts that fluoresce under UV light are considered viable (Figure 2B,C). Not all protoplasts that appear undamaged are still viable, and some viable protoplasts may be in the process of dying by evacuolation (Figure 2C). In these cells, the vacuole is swelling and will eventually be ejected from the cell membrane.
Figure 3 shows protoplasts transformed with pJT01, a 4.3 kb maize expression plasmid derived from CD3-911 (ABRC)15. Transfection results in the expression of sGFP tagged with a C-terminal nuclear localization signal from c-Myc (Figure 3A). The proportion of sGFP-expressing protoplasts was measured over multiple experiments using a hemocytometer. The median transfection efficiency achieved by this method was 40% after overnight incubation (n = 9, Figure 4, Table 1), which is comparable to previously published methods15. Depending on the experimental application, if the plasmid or plasmid library lacks a fluorescent reporter, it is recommended to include a positive control to confirm transfection.

Figure 1: Representative etiolated maize seedling grown in darkness for 10 days after germination. The arrows indicate the second and third leaves, which are best suited for protoplasting. Please click here to view a larger version of this figure.
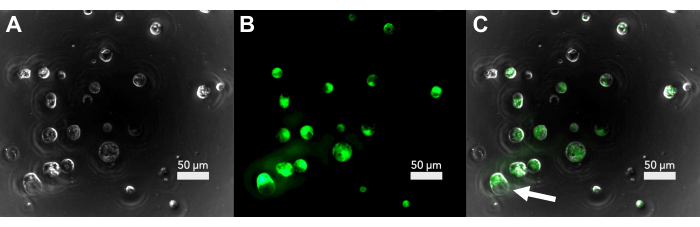
Figure 2: Fluorescence microscopy of the protoplast viability after isolation. (A) Brightfield image of protoplasts immediately after isolation. (B) The fluorescence signal of the FDA-stained protoplasts. (C) Overlap of brightfield and fluorescence showing the viable protoplasts. The arrow shows a protoplast undergoing evacuolation. Please click here to view a larger version of this figure.
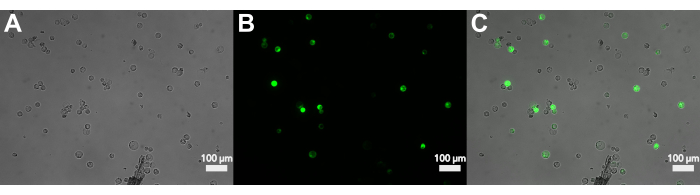
Figure 3: Fluorescence microscopy of protoplasts transfected with sGFP. (A) Brightfield image of protoplasts after transfection. (B) The fluorescence signal of the transfected protoplasts in the GFP channel. (C) Overlap of brightfield and fluorescence showing the transformed protoplasts. Please click here to view a larger version of this figure.
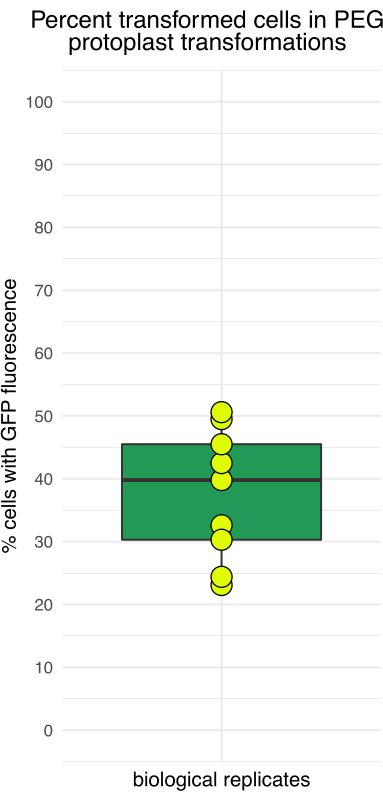
Figure 4: Percentage of protoplasts showing GFP fluorescence after 16 h of incubation. The transfection efficiency varied among independent trials, with the lowest efficiency around 20% and the highest close to 50%. Please click here to view a larger version of this figure.
| Trial | GFP cells counted | Total cells counted | GFP percentage |
| 1 | 104 | 261 | 39.8 |
| 2 | 148 | 298 | 49.5 |
| 3 | 84 | 258 | 32.6 |
| 4 | 137 | 271 | 50.6 |
| 5 | 127 | 299 | 42.5 |
| 6 | 107 | 235 | 45.5 |
| 7 | 83 | 360 | 23.1 |
| 8 | 134 | 549 | 24.4 |
| 9 | 61 | 201 | 30.3 |
| Mean | 109 | 304 | 36 |
| Standard deviation | 29 | 102 | 10 |
Table 1: Representative protoplast transfection efficiency. Data from nine recent protoplasting trials in the authors' laboratory.
Supplemental File 1: GenBank-formatted text file with the sequence of pJT01. A 4.3 kb sGFP expression plasmid serves as a positive control in transfected maize protoplasts. Please click here to download this File.
Discussion
As reported previously, the quality of plant material used for protoplasting is essential for obtaining high-quality protoplasts16,17,18. It is crucial to select healthy unblemished leaves. This work used the popular research maize cultivar B73. We have not tested other cultivars. The maize seedlings used for this protocol were grown in darkness because etiolated leaves produce protoplasts with greater viability 16 h after transfection compared to protoplasts from green leaves. For applications that do not require overnight incubation, using green or greening leaf material would be possible.
A critical step is properly cutting the leaf material into thin slices before enzymatic digestion. This process increases the surface area for the mesophyll cells to be exposed to the enzyme solution, which increases the number of recovered protoplasts. The distal leaf tips are discarded, and thin (≤1 mm) slices are cut with new razor blades, which are switched out immediately when they begin to dull. For a given amount of tissue, thinner slices yield more protoplasts provided the correct technique is employed to minimize the crushing of the leaves.
Washing the protoplasts correctly is also important, as they are quite delicate after undergoing cell wall removal. After digestion, all the solutions are kept on ice, and the dark-grown protoplasts are protected from light. The osmolarity and pH are buffered by performing the washes in MMG solution. To isolate the protoplasts from the less dense cellular debris, centrifugation takes place at 100 x g. Protoplasts centrifuged at 200 x g show similar viability after isolation but lower viability the following day, and they transform approximately half as well. We recommend checking the viability of the protoplasts by staining with FDA (fluorescein diacetate) immediately after isolation; the normal viability ranges between 70%-90%. Protoplasts with viability lower than 40% after isolation yield few transformants.
Pelleting and washing are easier when working with many protoplasts because a clearly visible pellet is obtained after transfection. For this reason, transfection is performed with 1 million or more protoplasts. When scaling up the protocol, an appropriately sized vessel should be selected for the incubation steps (1.5 mL, 15 mL, or 50 mL centrifuge tube), and the volume of solution used for the wash steps should be scaled accordingly. In any case, it is beneficial to centrifuge in ≤10 mL aliquots to ensure that the protoplasts do not remain in the supernatant. One innovation of this protocol is the removal of the 1 h incubation and wash step in W5 solution that is listed in other protocols14,19, which proved unnecessary in our hands.
As seen in other studies, the amount and quality of DNA are both critical for successful transfection19. For plasmids in the 4-6 kb range, 15 µg of DNA is used per 1 million protoplasts. Poor-quality DNA preparations can result in reduced viability after transfection, so we recommend using a commercial DNA extraction kit when isolating plasmids from bacteria. Incubating the protoplasts overnight is done at room temperature in the dark to allow for the transcription of the plasmid or plasmid library while minimizing stress. After overnight incubation, approximately half the number of originally transfected protoplasts is recovered. Protoplasts are diluted to a concentration of 500-1,000 cells/µL for overnight incubation. Protoplasts left to incubate overnight at concentrations higher than 1,000 cells/µL have lower recovery rates and lower viability. At higher concentrations, debris from damaged and dead protoplasts may contribute to the death of neighboring protoplasts. Washing the protoplasts after overnight incubation serves both to remove this debris and remove the excess plasmid.
Protocols such as these have the limitation that not all plant species or tissue types are amenable to protoplasting. Furthermore, gene expression differences may be induced by the protoplasting process.
In summary, we have detailed a simple and scalable protocol to isolate and transfect millions of viable protoplasts from the staple agricultural crop Z. mays. This method can transfect many more protoplasts using PEG, with a transformation efficiency nearly as high as electroporation-based methods15. The greater net number of transformants enables the testing of hundreds or thousands of constructs in large-scale experiments such as massively-parallel reporter assays12,13. Future genome-scale experiments in maize will allow scientists to better understand maize gene expression and gene regulation.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Human Genome Research Institute Interdisciplinary Training in Genomic Sciences (T32 training grant no. HG000035 to J.T.) and the National Institute of Health (NIGMS 1R35GM139532 to C.Q.) and by the National Science Foundation (RESEARCH-PGR IOS-1748843 and PlantSynBio 2240888 to CQ). We thank Andrea Gallovatti at Rutgers University for the gift of the maize seeds.
Materials
| 1.5 mL graduated microcentrifuge tubes | VWR | 490004-444 | |
| 15 mL Falcon conical centrifuge tube | Corning | 05-527-90 | |
| 250 mL Pyrex glass beaker | Fisher | 50-121-5005 | |
| 40 um nylon mesh cell strainer | Fisher | 22363547 | |
| 50 mL Falcon tube | Corning | 14-432-22 | |
| 72 cell propagation tray | T.O. Plastics | 725607C | |
| Aluminum foil | VWR | 89107-732 | |
| Bell jar | Bel-Art | F42022-0000 | |
| Benchtop centrifuge | Eppendorf | 5424-R | |
| Beta-mercaptoethanol | Sigma-Aldrich | M6250-250ML | Dissolve in ultrapure H2O to make a 1M stock solution. Store at room temperature. |
| Bovine serum albumin | Sigma-Aldrich | A2153 | |
| Calcium chloride dihydrate (CaCl2*2H2O) | PhytoTech Labs | C135 | Dissolve in ultrapure H2O to make a 1M stock solution. Store at room temperature. |
| Cellulase Onozuka R-10 | Duchefa Biochemie | C8001.0010 | |
| D-Mannitol | PhytoTech Labs | M562 | |
| Dissecting scope | Reichert | Microstar IV | |
| Fluorescein Diacetate (FDA) | Thermofischer Scientific | F1303 | Dissolve in acetone at 0.5% weight/volume to make a 1000X stock solution. Protect from light and store at -20 °C. |
| Fluorescence Illumination Systems | Prior | Lumen200 | |
| Fluorescence microscope | Leica | MDG36 | |
| High-capacity centrifuge | Eppendorf | 5810 R | |
| Macerozyme | Duchefa Biochemie | M8002.0005 | |
| Magnesium chloride (MgCl2) | Sigma-Aldrich | M292-1KG | Dissolve in ultrapure H2O to make a 1M stock solution. Store at room temperature. |
| Maize seeds | B73 | USDA-ARS | https://npgsweb.ars-grin.gov/gringlobal/accessiondetail?accid=PI+550473 |
| Medium binder clip | Uline | S-24649 | |
| MES | Sigma-Aldrich | M5287-250G | Dissolve in ultrapure H2O to make a 1M stock solution. Adjust pH to 5.7 using KOH. Protect from light and store at room temperature. |
| Micropipette 20-200 uL | Gilson | F123601G | |
| Nanodrop microvolume spectrophotometer | Thermofischer Scientific | ND-1000 | |
| NEB 5-alpha competent E. coli | New England BioLabs | C2987H | |
| Neubauer hemocytometer | Daigger Scientific | EF16034F | |
| No. 9 Single-edge razor blade | Excel | 20009 | |
| Orbital shaker | VWR | 89032-104 | |
| Poly(ethylene) glycol 4,000 | Sigma-Aldrich | 81240-1KG | |
| Potassium chloride (KCl) | Sigma-Aldrich | P9541-1KG | Dissolve in ultrapure H2O to make a 1M stock solution. Store at room temperature. |
| Professional Grade Vermiculite | NK Lawn & Garden | G208 | |
| Round-bottom glass centrifuge tube 30 mL | Kimble Chase | 45500-30 | |
| Rubber adapter for Corex centrifuge tubes | Corning | CLS8445AO | |
| Sphagnum peat moss | Premier Horticulture | 0128P | |
| TRIzol Reagent | Thermofischer Scientific | 15596018 | |
| Tygon vacuum tubing | VWR | 76336-844 | |
| Vacuum gauge | Wika | 4269978 |
References
- Jiang, F., Zhu, J., Liu, H. -. L. Protoplasts: A useful research system for plant cell biology, especially dedifferentiation. Protoplasma. 250 (6), 1231-1238 (2013).
- Wang, S., Tiwari, S. B., Hagen, G., Guilfoyle, T. J. AUXIN RESPONSE FACTOR7 restores the expression of auxin-responsive genes in mutant Arabidopsis leaf mesophyll protoplasts. The Plant Cell. 17 (7), 1979-1993 (2005).
- Hirner, A., et al. Arabidopsis LHT1 is a high-affinity transporter for cellular amino acid uptake in both root epidermis and leaf mesophyll. The Plant Cell. 18 (8), 1931-1946 (2006).
- Hwang, I., Sheen, J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 413 (6854), 383-389 (2001).
- Tan, M. -. L. M. C., Rietveld, E. M., van Marrewijk, G. A. M., Kool, A. J. Regeneration of leaf mesophyll protoplasts of tomato cultivars (L. esculentum): Factors important for efficient protoplast culture and plant regeneration. Plant Cell Reports. 6 (3), 172-175 (1987).
- Brandt, K. M., Gunn, H., Moretti, N., Zemetra, R. S. A streamlined protocol for wheat (Triticum aestivum) protoplast isolation and transformation with CRISPR-Cas ribonucleoprotein complexes. Frontiers in Plant Science. 11, 769 (2020).
- Nadakuduti, S. S., et al. Evaluation of methods to assess in vivo activity of engineered genome-editing nucleases in protoplasts. Frontiers in Plant Science. 10, 110 (2019).
- OECD, Food and Agriculture Organization of the United Nations. OECD-FAO Agricultural Outlook 2021-2030. OECD, Food and Agriculture Organization of the United Nations. , (2021).
- Gallois, P., Marinho, P. Leaf disk transformation using Agrobacterium tumefaciens-expression of heterologous genes in tobacco. Plant Gene Transfer and Expression Protocols. 49, 39-48 (1995).
- Schäffner, A. R., Sheen, J. Maize rbcS promoter activity depends on sequence elements not found in dicot rbcS promoters. The Plant Cell. 3 (9), 997-1012 (1991).
- Sheen, J. Molecular mechanisms underlying the differential expression of maize pyruvate, orthophosphate dikinase genes. The Plant Cell. 3 (3), 225-245 (1991).
- Jores, T., et al. Identification of plant enhancers and their constituent elements by STARR-seq in tobacco leaves. The Plant Cell. 32 (7), 2120-2131 (2020).
- Jores, T., et al. Synthetic promoter designs enabled by a comprehensive analysis of plant core promoters. Nature Plants. 7 (6), 842-855 (2021).
- Ricci, W. A., et al. Widespread long-range cis-regulatory elements in the maize genome. Nature Plants. 5 (12), 1237-1249 (2019).
- Sheen, J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiology. 127 (4), 1466-1475 (2001).
- Jeon, J. M., et al. Efficient transient expression and transformation of PEG-mediated gene uptake into mesophyll protoplasts of pepper (Capsicum annuum L.). Plant Cell, Tissue and Organ Culture. 88 (2), 225-232 (2007).
- Pindel, A. Optimization of isolation conditions of Cymbidium protoplasts. Folia Horticulturae. 19, 79-88 (2007).
- Ren, R., et al. Highly efficient leaf base protoplast isolation and transient expression systems for orchids and other important monocot crops. Frontiers in Plant Science. 12, 626015 (2021).
- Yoo, S. -. D., Cho, Y. -. H., Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nature Protocols. 2 (7), 1565-1572 (2007).

