Single Drosophila Ommatidium Dissection and Imaging
Summary
The limiting factor in the use of the adult Drosophila eye to study neurodegeneration and cell biology is the difficult imaging of intracellular processes. We describe the dissection of single ommatidia to generate a bona-fide primary neuronal cell culture, which can be subject to drug treatment and advanced imaging.
Abstract
The fruit fly Drosophila melanogaster has made invaluable contributions to neuroscience research and has been used widely as a model for neurodegenerative diseases because of its powerful genetics1. The fly eye in particular has been the organ of choice for neurodegeneration research, being the most accessible and life-dispensable part of the Drosophila nervous system. However the major caveat of intact eyes is the difficulty, because of the intense autofluorescence of the pigment, in imaging intracellular events, such as autophagy dynamics2, which are paramount to understanding of neurodegeneration.
We have recently used the dissection and culture of single ommatidia3 that has been essential for our understanding of autophagic dysfunctions in a fly model of Dentatorubro-Pallidoluysian Atrophy (DRPLA)3, 4.
We now report a comprehensive description of this technique (Fig. 1), adapted from electrophysiological studies5, which is likely to expand dramatically the possibility of fly models for neurodegeneration. This method can be adapted to image live subcellular events and to monitor effective drug administration onto photoreceptor cells (Fig. 2). If used in combination with mosaic techniques6-8, the responses of genetically different cells can be assayed in parallel (Fig. 2).
Protocol
1. Dissection of the Drosophila retina
- Fill a one well of a 3-Well Glass Slide with phosphate buffered saline (PBS 1X), and then anesthetize flies by CO2. Both males and females can be used for this experiment.
- Once the flies are anesthetized, pick up a single fly using forceps and drop it into the small dish of PBS.
- Under a dissecting scope, pinch the proboscis with forceps, and then detach the head from the body by severing the neck using a second set of forceps.
- Holding the fly head from the proboscis, cut in half along the left/right midline the fly head with microscissors to reveal the brain.
- Use a cut pipette tip to pick up the retina, and transfer to a second well of the 3-Well Glass Slide filled with Schneider medium.
- First, peel away most of the head cuticle and then remove the brain with forceps. Keep the retina sandwiched between the lamina and the cornea.
- Next, transfer the retina onto a drop (50 μl) of fresh Schnieder medium placed directly of the glass slide.
2. Dissection of the ommatidia
- Depending on the final purpose of the dissection (whether immediate imaging or culture) and on the optics of the fluorescent microscope (direct or inverted), this step can take place either on a microscope slide or in a 24-well plate. For the final purpose of this dissection, the ommatidia will be dissected directly onto the glass slide.
- Grasp either the most dorsal or ventral end of the retina with forceps. Using microscissors, cut the retina in half along the dorsal/ventral midline to expose the ommatidia in the middle of the eye.
- Continue to grasp the retina such that the cornea is facing down and the lamina is facing up. Using a fine tungsten needle, detach the ommatidia from the lamina and the cornea.
- Single ommatidia and groups of ommatidia will collect on the bottom of the well or slide. Remove any larger debris that may have collected before proceeding.
3. Treatment, staining and imaging
- To analyze autophagy, dilute 1:1000 Lysotracker in the Schneider drop (first dilute 1:10 in Schneider and then add 0.5 μl to the drop on the slide), mix and wait 10 seconds.
- Remove the excess liquid from the slide, being careful to leave the ommatidia behind. This is done best with a fine gel loading tip.
- Add a drop of Vectashield to cover the ommatidia, apply a coverslip and seal with nailpolish.
4. Representative Results:
The good execution of the protocol should leave a number of single ommatidia and of small groups of ommatidia kept together by fragments of lamina but nicely separated on the corneal side. These can be imaged as in this example to visualize Atg8:GFP and Lysotracker, showing autophagosomes, lysosomes and autophagolysosomes (Fig. 3). The method can be used for live imaging, however in this case it is to be noted that autofluorescence of the Schneider’s medium will make it difficult to distinguish low intensity fluorescent signals. An additional issue is that the pigment granules that remain attached to the photoreceptors are intensely fluorescent, especially in the red channel. A brightfield picture may be required to distinguish them from genuine organelles.
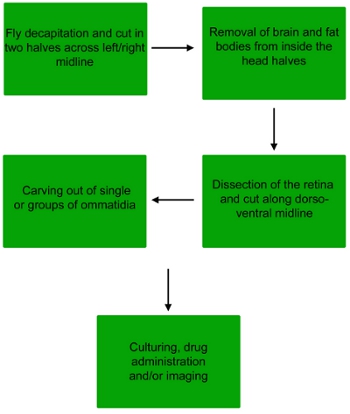
Figure 1. Flow chart of the dissection procedure to obtain single ommatidia or small groups of ommatidia from the fly retina.
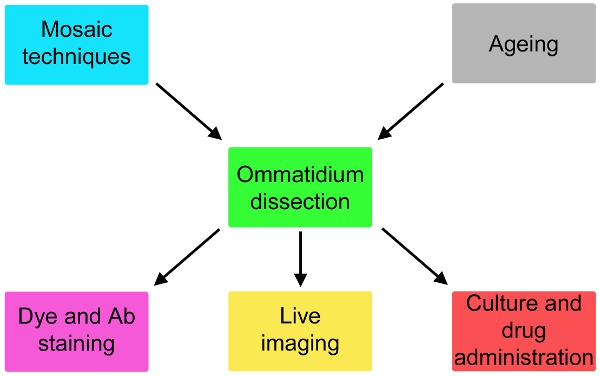
Figure 2. Possible variations of the simple protocol here describe that involve fly genetics and ageing upstream of the dissection and staining or culturing for up to 24 hours and drug administration downstream of the dissection.
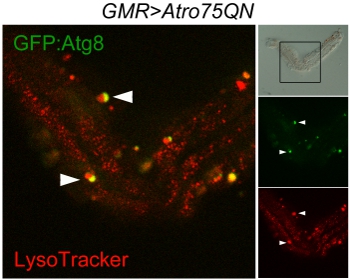
Figure 3. Confocal scan of autophagosomes and lysosomes in a single ommatidium dissected from a w; GMR-Gal4, UAS-Atro75QN; UAS-GFP::Atg8/+ fly, expressing a polyglutamine Atrophin mutant and aged 12 days at 29°C. The brightfield panel (top right) displays the almost intact structure of the ommatidium and the frame indicates the scanned area. Red marks Lysosomes, green the autophagosomes, auto-lysosomes, resulting by the fusion of the two organelles, appear yellow. The arrowheads indicate two ongoing fusion events between autophagosomes and lysosomes.
Discussion
The single ommatidium dissection presented here enables collection of deeper cell biological information about processes like neurodegeneration, when modeled in the Drosophila eye. The photoreceptor neurons are more easily accessible than other neurons and they cytoplasm is particularly large, and thus suitable to study vesicle dynamics, in the very same cells used proficiently in many models to quantify neurodegeneration in vivo. The critical aspect of the dissection is mainly the ability to perform it on unfixed tissue that must never be exposed to air or dry up. The possibilities for expansion of this basic technique are such (Fig. 2) that it may as well revolutionise the information obtained from eye models of neurodegeneration, allowing all kinds of manipulations possible in a primary neuronal culture coupled to the amazing genetic modifications possible in flies.
In combination with mosaic techniques, extremely useful in neurodegeneration studies9 it may allow identification of cell biological alterations in a specific subset of mutant ommatidia, in presence of wt controls from the very same fly.
When dissected ommatidia are kept in culture, reponses to drugs like rapamycin10 or parquat11 may be better controlled and quantified than when administered to living flies. The main limitation of the system is constituted by the survival time of the dissected ommatidia in culture, which we have not been able to expand much beyond 24 hours.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Bernard Charroux for discussions. This work was funded by the KCL Biomedical School.
Materials
| Name of the reagent | Company | Catalogue number | Comments (optional) |
|---|---|---|---|
| Portable CO2 Dispenser | Flystuff | 59-150 | Refills also available |
| DUMONT Tweezer No 5 | AGAR Scientific | T5034 | |
| 3-Well Glass Slide | EMS | 71561-01 | |
| Micro scissors | VWR | HAMMHSB516-09 | |
| Schneider’s Medium | VWR | 733-1663 | |
| LysoTracker Red DND-99 | Invitrogen | L7528 | |
| Superfrost Slides | VWR | 631-0108 | |
| Vectashield with Dapi | Vector Labs | H-1200 | |
| Coverslips | VWR | 631-1336 |
References
- Marsh, J. L., Thompson, L. M. Drosophila in the study of neurodegenerative disease. Neuron. 52, 169-178 (2006).
- Mizushima, N., Levine, B., Cuervo, A. M., Klionsky, D. J. Autophagy fights disease through cellular self-digestion. Nature. 451, 1069-1075 (2008).
- Nisoli, I. Neurodegeneration by polyglutamine Atrophin is not rescued by induction of autophagy. Cell Death Differ. 17, 1577-1587 (2010).
- Charroux, B., Fanto, M. The fine line between waste disposal and recycling: DRPLA fly models illustrate the importance of completing the autophagy cycle for rescuing neurodegeneration. Autophagy. 6, 667-669 (2010).
- Hardie, R. C. Voltage-sensitive potassium channels in Drosophila photoreceptors. J Neurosci. 11, 3079-3095 (1991).
- Lee, T., Luo, L. Mosaic analysis with a repressible cell marker (MARCM) for Drosophila neural development. Trends in neurosciences. 24, 251-254 (2001).
- Xu, T., Rubin, G. M. Analysis of genetic mosaics in developing and adult Drosophila tissues. Development. 117, 1223-1237 (1993).
- Gambis, A., Dourlen, P., Steller, H., Mollereau, B. Two-color in vivo imaging of photoreceptor apoptosis and development in Drosophila. Dev Biol. 351, 128-1234 (2011).
- Napoletano, F. Polyglutamine Atrophin provokes neurodegeneration in Drosophila by repressing fat. The EMBO journal. 30, 945-958 (2011).
- Ravikumar, B. Inhibition of mTOR induces autophagy and reduces toxicity of polyglutamine expansions in fly and mouse models of Huntington disease. Nature. 36, 585-595 (2004).
- Zou, S., Meadows, S., Sharp, L., Jan, L. Y., Jan, Y. N. Genome-wide study of aging and oxidative stress response in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America. 97, 13726-13731 (2000).

