Summary
This video demonstrates three types of morphometric analyses of the retina, which include measuring the inner nuclear layer thickness, quantifying the number of retinal ganglion cells (RGCs) and measuring the sizes of RGCs. The technique can offer a simple but scientific platform for morphometric analyses.
Abstract
Morphometric analyses of retinal sections have been used in examining retinal diseases. For examples, neuronal cells were significantly lost in the retinal ganglion cell layer (RGCL) in rat models with N-methyl-D-aspartate (NMDA)–induced excitotoxicity1, retinal ischemia-reperfusion injury2 and glaucoma3. Reduction of INL and inner plexiform layer (IPL) thicknesses were reversed with citicoline treatment in rats’ eyes subjected to kainic acid-mediated glutamate excitotoxicity4. Alteration of RGC density and soma sizes were observed with different drug treatments in eyes with elevated intraocular pressure3,5,6. Therefore, having objective methods of analyzing the retinal morphometries may be of great significance in evaluating retinal pathologies and the effectiveness of therapeutic strategies.
The retinal structure is multi-layers and several different kinds of neurons exist in the retina. The morphometric parameters of retina such as cell number, cell size and thickness of different layers are more complex than the cell culture system. Early on, these parameters can be detected using other commercial imaging software. The values are normally of relative value, and changing to the precise value may need further accurate calculation. Also, the tracing of the cell size and morphology may not be accurate and sensitive enough for statistic analysis, especially in the chronic glaucoma model. The measurements used in this protocol provided a more precise and easy way. And the absolute length of the line and size of the cell can be reported directly and easy to be copied to other files. For example, we traced the margin of the inner and outer most nuclei in the INL and formed a line then using the software to draw a 90 degree angle to measure the thickness. While without the help of the software, the line maybe oblique and the changing of retinal thickness may not be repeatable among individual observers. In addition, the number and density of RGCs can also be quantified. This protocol successfully decreases the variability in quantitating features of the retina, increases the sensitivity in detecting minimal changes.
This video will demonstrate three types of morphometric analyses of the retinal sections. They include measuring the INL thickness, quantifying the number of RGCs and measuring the sizes of RGCs in absolute value. These three analyses are carried out with Stereo Investigator (MBF Bioscience — MicroBrightField, Inc.). The technique can offer a simple but scientific platform for morphometric analyses.
Protocol
1. Tools
Microscope, Nikon
Stereo Investigator, MBF Bioscience – MicroBrightField, Inc.
2. Preparation
- Before working on any morphometric analysis, each retina sample is sectioned at 4 micron thickness and undergoes the H&E staining.
- The retina section is divided into 4 regions – upper peripheral region, the upper central region, the lower peripheral region, and the lower central region.
- After defining the regions, an image of each region is acquired at 10×40 magnifications under the bright beam microscope of Nikon.
- Now, we are ready to analyze morphometry of the retina.
3. Measuring INL thickness
- Four measurements of INL thickness in each region were measured. Four lines will be drawn perpendicularly from the contour baseline to the outer border of the INL.
- Open the software called Stereo Investigator and choose the 40x Lens from the Main Toolbar.
- Select “Image Open” under the tab “File”. Choose an image of a desired retinal region that you would like to analyze.
- Choose the type of contour line from the Dropdown list box of the Main Toolbar and press the “Auto Move” button.
- Right click in the tracing window, and select “Simple Click Tracing”.
- Draw the contour line along the inner border of the INL.
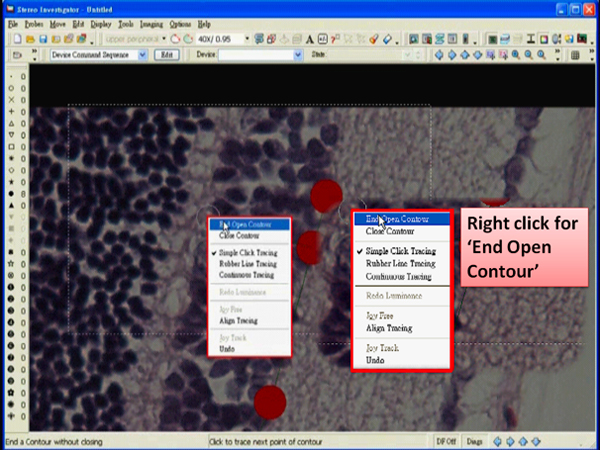
- When the drawing is done, right click in the tracing window and choose “End Open Contour”.
- Select a marker at the marker toolbar and add markers at the junction points.
- After adding all the markers, deselect the marker icon at the Marker Toolbar.
- Position a cursor at the centre of one of the markers.
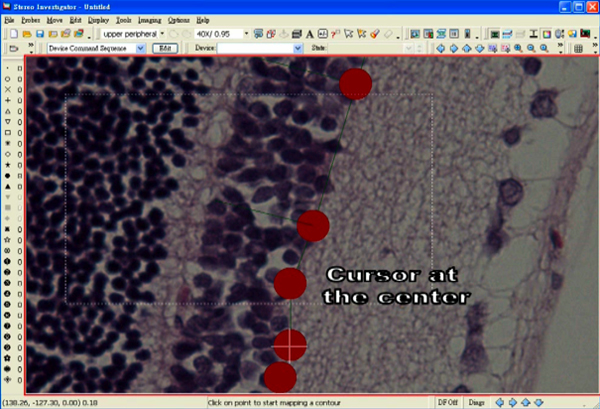
- Select “Quick Measure Angle” under the tab “Tools”.
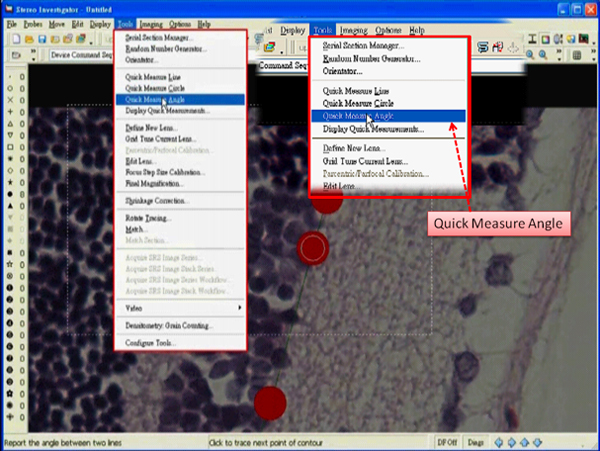
- Right click in the tracing window and deselect the “Continuous” option.
- Measure an angle by first clicking at the center of a marker, which is adjacent to the marker selected for an end of the line segment.
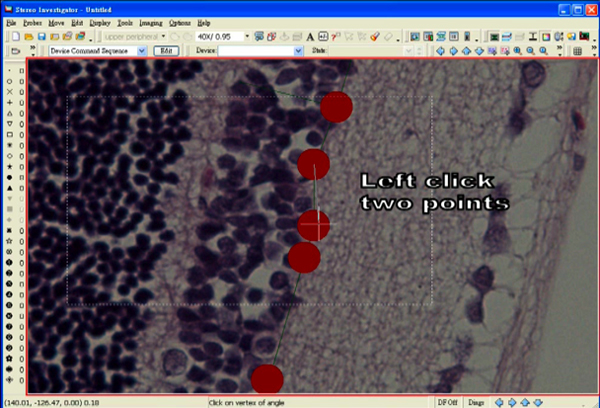
- Then, move the mouse to the center of the concerned marker and click again.
- Extend the rubber band line to the outer border of the INL.
- Adjust the angle until 90° is reached.
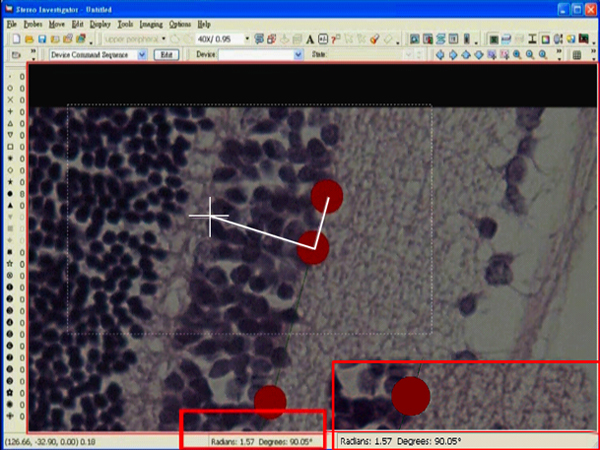
- Left click at the desired position. A dialog box showing an angle measurement will pop up right after the click.
- Press the “Enter” key while holding the mouse still.
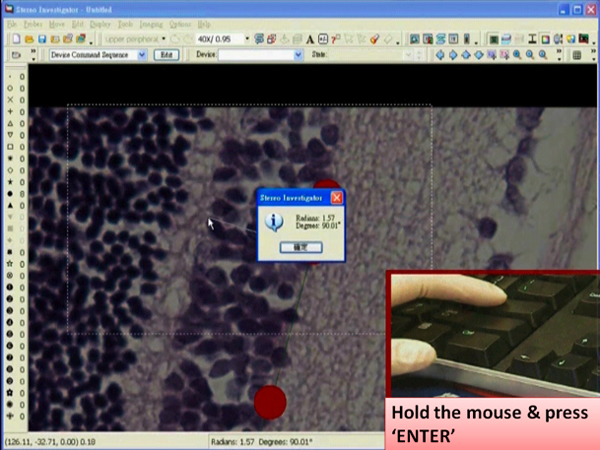
- Left click the tracing window so that a line is drawn from one end to the new selected end.
- Right click in the tracing window and choose “End Open Contour”.
- Draw the other lines at different positions in exactly the same way as described before.
- After drawing all the lines, select the Editing mode and rename the 4 line segments accordingly.
- Four measurements can be obtained by pressing “Contour Measurement” button.
- Select these 4 measurements and paste them to a spreadsheet by clicking “Copy to Clipboard”.
- Select “Save As” under the tab “File”. The tracing data can be saved as a data file.
4. Quantifying the number of RGCs
- Select “Image Open” under the tab “File”. Choose an image of a desired retinal region that you would like to analyze.
- Choose the type of contour line from the Dropdown list box of the Main Toolbar.
- Draw a contour line along the border of nerve fiber layer (NFL).
- End the tracing with the option “End Open Contour”.
- At each end, add one marker at the tip of the line and another at the position near the tip.
- Deselect the marker icon at the marker toolbar.
- Position a cursor at the centre of the markers at the tip.
- Select “Quick Measure Angle” under the tab “Tools”.
- Right click in the tracing window and deselect the “Continuous” option.
- Adjust the rubber band lines and measure an angle of 90°.
- Press the “Enter” key after a dialog box is popped up.
- Left click the tracing window so that a new line can be perpendicularly extended from the tip.
- Draw another new line in the exactly the same way at another end.
- Two boundaries are created within which the number of RGCs is counted.
- Choose one boundary as an exclusion line which the cell touches is not counted.
- Select one type of marker and attach one to each live cell.
- Select another type of marker for the dead cells.
- The number of each type of markers placed will be displayed alongside the marker icon accordingly at the marker toolbar.
- Select the Editing mode and pick up the contour baseline. Right click in the tracing window and rename the line.
- The length of the contour baseline can be obtained by pressing “Contour Measurement” button.
- The number of the cells can then be expressed per mm of the length.
- Select “Save As” under the tab “File”. The tracing and the marker data can be saved as a data file.
5. Measuring the cell size of a RGC cell
- Select “Image Open” under the tab “File”. Choose an image of a desired retinal region that you would like to analyze.
- Zoom in a live cell and choose the type of contour line from the Dropdown list box of the Main Toolbar.
- Draw a contour line along the circumference of the cell.
- At about the end, right click the tracing window and select “End close contour”.
- The area of the cell will be displayed under the item of Area in the Contour Measurement dialog box.
Discussion
1. How to obtain a more accurate thickness measurement?
You can enlarge the image by clicking “Zoom in” button and left click in the tracing window. The INL border can be seen clearly. If the tip of the line is not at the outer border of the INL, we can adjust the line under the Editing mode. After picking the concerned line, right click the tracing window and choose “Insert Point in Selected Contour”. A point can be added at the outer border while the point added should be along the same line. Then, you have to delete the original point and a line with a more accurate length can be obtained.
2. How to maintain 90° of the rubber band line?
After a dialog box showing the degree is popped up, you have to hold the mouse still while pressing the “Enter” key. Then, left click the tracing window so that a new line can be drawn.
3. How do live cells and dead cells look like?
Those cells in a roughly round shape with cytoplasm and nucleus clearly observed are considered as live RGCs. Those comparably small and condensed cells are considered as dead cells.
4. Those retina samples were undergone H&E staining in the current protocol. Is it possible to measure the retina sample with fluorescent staining?
Yes. Although current protocol is on the H&E staining retinal sections, the retinal sample with fluorescent staining can also be measure the same way. For example, as shown in Figure 1 using DAPI staining, all the nuclei will be stained and visualized blue in color under a 405 nm filter. Nuclei in the retinal ganglion cell layer (RGCL), inner nuclear layer (INL) and outer nuclear layer (ONL) can be seen. Then the thickness of different layers can be detected in a similar way demonstrated in this video.

Figure 1.
About the size of different stained fluorescent signals, our group has published a paper concerning about the size of neurons in the RGCL in a rat glaucoma model (Luo et al, 2009).
Disclosures
The authors have nothing to disclose.
Acknowledgements
The work on eye research in this laboratory is supported by American Health Assistant Foundation, Azalea (1972) Endowment Fund, and HKU Small Project Fund (20097176185).
Materials
| Name of the reagent | Type | Company | Catalogue number |
| Stereo Investigator | MBF Bioscience Analysis Software | MicroBright Field | |
| Microscope | Olympus BX51 | Olympus Corporation | BX51 |
References
- Lam, T. T., Abler, A. S., Tso, M. O. N-Methyl-D-Aspartate (NMDA)–Induced Apoptosis in Rat Retina. Investigative Ophthalmology & Visual Science. 40, 2391-2397 (1999).
- Lam, T. T., Abler, A. S., Tso, M. O. Apoptosis and caspases after ischemia-reperfusion injury in rat retina. Invest. Ophthalmol. Vis. Sci. 40, 967-975 (1999).
- Luo, X. G., Chiu, K., Lau, H. S., Lee, V. W. H., Yung, K. K. L., So, K. F. The Selective Vulnerability of Retinal Ganglion Cells in Rat Chronic Ocular Hypertension Model at Early Phase. Cellular and Molecular Neurobiology. 29 (8), 1143-1151 (2009).
- Han, Y. S., Chung, I. Y., Park, J. M., Yu, J. M. Neuroprotective effect of citicoline on retinal cell damage induced by kainic acid in rats. Korean J. Ophthalmol. 19, 219-226 (2005).
- Hernandez, M., Urcola, J. H. Retinal ganglion cell neuroprotection in a rat model of glaucoma following brimonidine, latanoprost or combined treatments. Exp. Eye Res. 86, 798-806 (2008).
- Chan, H. C., Chang, R. C. C., Ip, A. K. C., Chiu, K., Yuen, W. H., Zee, S. Y., So, K. F. Neuroprotective effects of Lycium barbarum Lynn on protecting retinal ganglion cells in an ocular hypertension model of glaucoma. Experimental Neurology. 203, 269-273 (2007).

