A Duplex Digital PCR Assay for Simultaneous Quantification of the Enterococcus spp. and the Human Fecal-associated HF183 Marker in Waters
Summary
This manuscript describes a duplex digital PCR assay that can be used to simultaneously quantify Enterococcus spp. and the HF183 genetic markers as indicators of general and human-associated fecal contamination in recreational waters.
Abstract
This manuscript describes a duplex digital PCR assay (EntHF183 dPCR) for simultaneous quantification of Enterococcus spp. and the human fecal-associated HF183 marker. The EntHF183 duplex dPCR (referred as EntHF183 dPCR hereon) assay uses the same primer and probe sequences as its published individual quantitative PCR (qPCR) counterparts. Likewise, the same water filtration and DNA extraction procedures as performed prior to qPCR are followed prior to running dPCR. However, the duplex dPCR assay has several advantages over the qPCR assays. Most important, the dPCR assay eliminates the need for running a standard curve and hence, the associated bias and variability, by direct quantification of its targets. In addition, while duplexing (i.e. simultaneous quantification) Enterococcus and HF183 in qPCR often leads to severe underestimation of the less abundant target in a sample, dPCR provides consistent quantification of both targets, whether quantified individually or simultaneously in the same reaction. The dPCR assay is also able to tolerate PCR inhibitor concentrations that are one to two orders of magnitude higher than those tolerated by qPCR. These advantages make the EntHF183 dPCR assay particularly attractive because it simultaneously provides accurate and repeatable information on both general and human-associated fecal contamination in environmental waters without the need to run two separate qPCR assays. Despite its advantages over qPCR, the upper quantification limit of the dPCR assay with currently available instrumentation is approximately four orders of magnitude lower than that achievable by qPCR. Consequently, dilution is needed for measurement of high concentrations of target organisms such as those typically observed following sewage spills.
Introduction
Quantitative PCR (qPCR) methods have been widely used for recreational water quality monitoring and microbial source tracking applications because they are faster, more flexible and specific compared to traditional culture-based methods. Consequently, qPCR is recommended for achieving rapid water monitoring results for general fecal indicators such as Enterococcus spp. in the revised USEPA recreational water quality criteria1. Many qPCR assays have also been developed and validated for identifying sources of fecal contamination in environmental waters2. Among these, the HF183 marker assays are among the most frequently used for identifying human-associated fecal contamination3.
However, qPCR results can often be biased because they rely on standard curves for quantification. qPCR quantifies unknown concentrations of target in samples by interpolating their quantification cycle (Cq) from a standard curve that describes a linear relationship between Cq and logarithmic quantity of a set of serially-diluted standards. Lack of reliable and consistent standard reference material can therefore greatly bias qPCR results. Studies showed that qPCR results differed by approximately half a log using standards from different vendors4 and by 2-fold using different batches of standards from the same vendor5.
Digital PCR (dPCR) technology quantifies an unknown sample by counting the frequency of positives in large numbers of miniature PCRs that are generated by partitioning a bulk qPCR into thousands or millions of uniform nanoliter or picoliter reactions6. It is referred as droplet digital PCR (i.e., this article) or chamber digital PCR, respectively, if the partitions are water-in-oil droplets (i.e., this article) or small chambers on a chip. A higher sample concentration will result in presence of target DNA (hence positive PCR) in a higher proportion of partitions, a relationship approximated by Poisson distribution. As such, the binary results (positive or negative PCR) are recorded and the frequency of positive droplets is used to calculate unknown sample concentrations directly via Poisson approximation, eliminating the need for a standard curve as in qPCR.
This binary nature of digital PCR quantification provides additional advantages over qPCR7. Because delayed amplification can still yield a positive PCR, dPCR quantification is more robust against inhibition that reduces amplification efficiency. Similarly, dPCR quantification is not affected by variability in Cq values as is qPCR, leading to higher precision of dPCR compared to qPCR8-10.
In addition, the partitioning process ensures a very small amount of target DNA present in each droplet, effectively minimizing substrate competition (during amplification of different DNA targets) that are common obstacles to achieving duplexing (i.e. an assay simultaneously measures two DNA targets) in qPCR. As such, whether run in duplex or simplex (i.e. one target is measured in a single assay) dPCR produces nearly indistinguishable quantification of both targets7,11.
The goal of the digital PCR assay (EntHF183 dPCR) described in this article is to obtain simultaneous direct quantification of the general fecal indicator Enterococcus spp. and the human-fecal associated HF183 marker with improved precision, reproducibility and robustness against inhibition compared to its qPCR counterparts. This assay has been successfully used for quantifying fecal contamination in environmental freshwater, marine water, and various fecal material7, but can also be applied to any other types of waters and wastewaters. This assay holds great promise for analyzing sediments or soils due to its high tolerance to inhibition, but further testing is required because of complexities of inhibitor mixes in these types of samples.
Protocol
1. Assay Mixture Preparation
- Make 100 µmol/L stock concentrations for all primers in molecular grade water and probes in TE pH 8 buffer [Entero F1A, Entero R1, GPL813TQ 12; HF183-1, BthetR1, BthetP13].
Note: Probes for the two targets are fluorescently labeled with different fluorophores7 as indicated in List of Materials/Equipment. - Prepare master mix by mixing, per reaction planned, 12 µl digital PCR mix (2x stock, see List of Materials/Equipment), 0.216 µl each forward and reverse primer, 0.06 µl each fluorescent probe, and 5.016 µl nuclease free water (final concentration: 900 nM each primer, 250 nM each probe). Pipette up and down at least 10 times to mix while taking caution not to introduce air bubbles in the solution.
- Pipette 18 µl master mix (from step 1.2) into a regular PCR tube or plate, mixing in 6 µl DNA template (extracted as described previously13) to make assay mixture for droplet generation. If running samples in duplicate, pipet 36 µl of master mix and 12 µl DNA template into each well. Leave the corresponding replicate wells empty on the plate. Include positive controls (see List of Materials/Equipment) and no template controls (NTC) (i.e. with molecular grade water used as the template).
Note: Positive control is necessary to ensure the assay is running properly and the NTC is necessary to ensure there is no contamination within the plate and to set the fluorescent baseline later in data analysis. First time users are recommended to use both undiluted and diluted DNA samples.
2. Droplet Generation and PCR Plate Setup
- Mix the assay mixtures by pipetting up down approximately 15 times using a multichannel pipet. Ensure that the pipet tip stays within liquid to avoid making excess bubbles within the mixture.
- Insert cartridge 1 (containing 8 wells) into a white cartridge holder and click the cartridge holder shut. Cartridge 1 is now firmly in place and cannot be dislodged from the holder while generating droplets. Using a multichannel pipet, gently transfer 20 µl of assay mixture into the middle position of the cartridge marked 'Sample' without introducing air bubbles.
- Pipet in 70 µl of droplet generation oil to the left side of the cartridge (marked 'Oil').
- Cover cartridge with a gasket making sure the gasket is flat and held evenly by the 4 ticks toward the edge of the cartridge. Press the green-lit button on the droplet generator to open and place the cartridge. Press green-lit button again to close the generator.
Note: Once the door closes, the button is dimmed, the door cannot be reopened and droplet generation begins immediately continuing for approximately 1 min. - If doing more than 8 reactions, place cartridge 2 in a second white cartridge holder and prepare in the same manner as cartridge 1 while the droplet generation is in progress to save total setup time.
- Open the droplet generator door when the dim lit button turns green indicating completion of droplet generation. Remove the white cartridge holder containing cartridge 1, set aside, and place cartridge 2 onto the droplet generator.
- Remove the gasket from cartridge 1 and discard. Gently transfer the total volume (approximately 40 µl) of generated droplets from the third column of the cartridge (marked 'Droplets') onto a final PCR plate (maintained at room temperature) for thermal cycling.
Note: Do not unclick the white cartridge holder as the action may break the newly generated droplets; only open the cartridge holder after the droplets have been transferred to the final plate. - Repeat steps 2.1-2.7 for additional samples.
- When all the droplets are in the final PCR plate, place a pierceable foil cover on top of the plate and place it on a plate sealer. Set the sealer to 180 °C, press 'Play' on the sealer and let seal for 10 sec.
3. Thermal Cycling and Droplet Reading
- Place the sealed plate on the thermal cycler, one compatible with the final PCR plate used in step 2.7, and a temperature ramping speed of 2.0 °C/sec.
- Run thermal program as follows: 10 min at 95 °C, followed by 40 cycles of 30 sec at 94 °C and 60 sec at 60 °C, followed by a 10 min hold at 98 °C (optional: final hold at 4 or 15 °C).
- Upon completion of cycling, transfer the plate to a Droplet Reader for automatic measurement of fluorescence in each droplet in each well. Alternatively, store the plate at 4 °C for up to 3 days before droplet reading. Ensure the droplets are at room temperature before proceeding with reading.
- Open the accompanying software to setup the droplet reading. In the default 'Setup' menu containing a schematic of an empty 96 well plate, double click on well A1 to open the menu containing three sections: 'Sample,' 'Assay 1' and 'Assay 2.'
- In the 'Sample' section type the sample ID into the box labeled 'Name' and press enter or check the box to the right marked 'Apply.' Next, click the drop down menu labeled 'Experiment' and choose 'RED' (rare event detection) and click enter.
- Move to the section denoted 'Assay 1.' In the 'Name' section fill out the assay (e.g. Enterococcus) and click enter. In the box below labeled 'Type' click the drop down menu and choose 'Channel 1 Unknown' and click enter.
- Move to the section denoted 'Assay 2.' In the 'Name' section fill out the assay (e.g. HF183) and click enter. In the box below labeled 'Type' click the drop down menu and choose 'Channel 2 Unknown' and click enter. All the information from Steps 3.3.2 to 3.3.4 is now present in well A1.
- Name all subsequent wells containing droplets. To save total setup time, click 'Shift' or 'Ctrl,' to choose multiple wells simultaneously.
- When the digital depiction of the plate mirrors the physical plate, press 'OK' at the bottom right of the menu. In the new menu that appears at the top of the plate schematic, under the 'Template' section choose 'Save As' and name and save the plate.
- To the left of the screen click 'Run' and select appropriate Dye Set in the pop-out “Run Options” window. Data collection initiates and is displayed in real time in the software.
4. Data Analysis and Reporting
- When the reader is finished and a box appears stating 'Run Complete', click 'OK'.
Note: The software displays the final data file under the 'Analyze' section of the software. In this view, the software has all the wells in the plate selected and defaults to the '2D Amplitude' data plot. - Check the separation between the positive and negative droplets. Ensure that the fluorescence in all droplets in the NTC wells are near baseline.
- Click on the button labeled 'Events.' To the right of the presented histogram click the box named ‘Total’ and the box named “single” at the bottom to display the total number of accepted droplets per well. Exclude any wells containing <10,000 droplets7 by holding the Ctrl key and clicking on the well(s) to be excluded.
- Click on the button denoted as '1D Amplitude' to set the fluorescent threshold at approximately one standard deviation (500-700 fluorescence units) above the negative droplets in NTC wells for both targets. At the very left of the screen under the 'Auto Analyze' showing two threshold buttons, choose the icon to the right that has a solid pink horizontal line running through it.
- Click in the box to the left of 'Set Threshold' under each of the amplitude graphs and enter the appropriate fluorescence threshold values. The target concentrations in copy per µl reaction are then automatically calculated.
Note: The thresholds do not need to be identical for the different targets. - Export the results in a .csv file by clicking the 'Export' button in the upper left hand corner on the 1D Amplitude screen. In the .csv file, multiply the exported target concentration by 4 to convert it from copy of target (23S gene of Enterococcus spp. or the HF183 marker) per µl reaction to copy of target per µl DNA template.
Representative Results
A good EntHF183 duplex dPCR run should result in relatively high number of accepted droplets (ca. 10,000-17,000) and relatively large difference (ca. 5,000) between fluorescence values for negative and positive droplets7. Unusually low number of droplets likely indicates issues in the droplet generation process, and a cutoff of 10,000 droplets is suggested based on empirical data from the droplet dPCR system used in this article. A weaker separation (i.e. smaller fluorescence difference) between positive and negative droplets may indicate inhibition or degraded probes7.
Overall, the EntHF183 duplex dPCR assay produces highly comparable results to that from simplex qPCR when qPCR is corrected for biases associated with standards and there is no inhibition7. Results from the EntHF183 duplex dPCR assay and the corresponding simplex dPCR assays are highly consistent and often indistinguishable between the two formats7 (Figure 1). Additionally, the dPCR assay can also tolerate inhibitor concentrations one to two orders of magnitude higher than that tolerated by its qPCR counterparts7 (Figure 2).
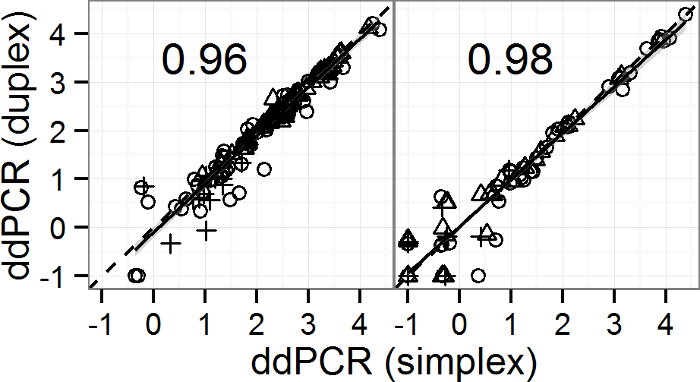
Figure 1: Quantification of fecal and water samples by the EntHF183 duplex dPCR assay and the corresponding simplex dPCR assays. Left and right panels display Enterococcus and HF183 quantification, respectively, with the corresponding correlation coefficients (p <0.001) between duplex and simplex results. Solid lines indicate the regression lines and the gray shading indicates the corresponding standard errors. Symbols indicate different types of samples (Circles, triangles and crosses denote fecal, freshwater, and marine water samples, respectively). Please click here to view a larger version of this figure.
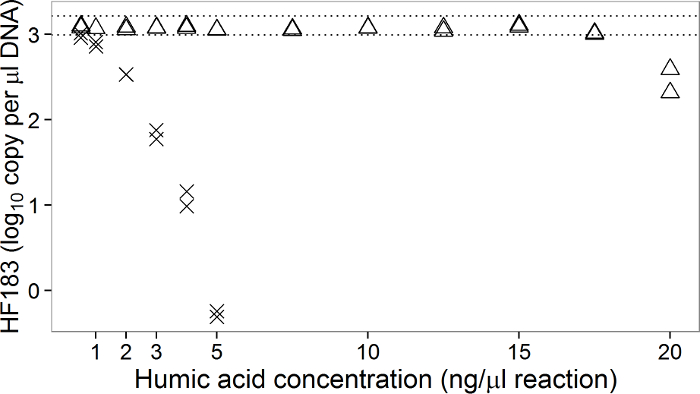
Figure 2: Simplex qPCR and duplex dPCR quantification of the HF183 marker in clean sewage DNA spiked with increasing concentrations of PCR inhibitors (humic acid). Triangles and x-crosses denote dPCR and qPCR quantification, respectively. The expected HF183 quantification in absence of inhibitors is defined as 95% confidence interval (i.e., between the two horizontal dotted lines). A "-1" result indicates non-detection. Enterococcus quantification shows similarly higher tolerance to inhibitors by the EntHF183 duplex dPCR assay than by the corresponding qPCR assay7. Please click here to view a larger version of this figure.
Discussion
There are a few critical steps in the protocol. First, mixing of the assay mixtures can be difficult because the master mix is more viscous than conventional qPCR master mixes. Simple vortexing may lead to insufficient mixing, which in turn leads to non-uniform distribution of DNA templates in the assay mixtures and subsequently in the droplets. The mixing-by-pipetting technique described in the protocol must be followed to ensure accurate dPCR quantification. Second, it is important to ensure droplets are generated at a uniform shape and size. Be mindful of occasions when the time the droplet generator takes to complete droplet generation on one cartridge is shorter than usual. This may indicate suboptimal droplet generation. If necessary, the droplet generation time on each cartridge can be recorded for troubleshooting. Third, it is important to transfer the entire 40 µl of generated droplets from the cartridge to the final PCR plate without shearing the droplets and avoid not having enough droplets for quantification. The following techniques may be used for the transfer: 1) Set the multichannel pipet to 40 µl and insert the tips into the right section of the cartridge (marked 'Droplets') at a 45° angle; 2) Pipet the droplets out slowly and ensure that all the droplets are pipetted up (take note if any droplets are left behind and need to be pipetted up for a second round); 3) Once the droplets are removed, transfer them to the final plate by putting the pipet tip against the well wall approximately half way down and expel the droplets slowly.
The EntHF183 duplex dPCR assay has great advantages over the existing individual qPCR assays for the same analytes. First, the duplex dPCR assay does not need standard curves for quantifying unknowns, thereby eliminating qPCR quantification biases associated with variability in standards7. Second, the duplex dPCR assay can tolerate inhibitor concentrations that are one to two orders of magnitude higher than that tolerated by the corresponding qPCR assays7. This feature makes the duplex dPCR assay very attractive for analyzing environmental samples (e.g. stormwater samples) that often contain substances inhibitory to PCR. Third, the capacity to simultaneously quantify both Enterococcus spp. and the HF183 marker in one duplex dPCR assay (vs. having to quantify them separately in two qPCR assays) is more cost-effective7 and improves data quantity by avoiding accumulative pipetting variability associated with conducting two separate assays (vs. one duplex dPCR assay). Fourth, the duplex dPCR assay also demonstrated higher precision and repeatability compared to the corresponding qPCR assays, making the former a good alternative in comparative studies7.
Nevertheless, limitations exist for this method (advantages and limitations described in detail elsewhere7). First, the number of accepted droplets per reaction determines the upper quantification limit (UQL) of dPCR. The dPCR instrument used in this work (refer to the List of Materials/Equipment) has been reported to have a UQL of 105 copy per reaction. This corresponds to 5 x 105 Enterococcus cells and 2 x 106 HF183 copies per 100 ml of water assuming the previously7 described DNA extraction protocol is followed and 100 µl DNA is obtained (i.e. per the elution step during the DNA extraction) from 100 ml of water without any loss during sample processing prior to dPCR7. Environmental waters generally do not exceed such concentrations of Enterococcus and HF183. However, sample dilution should be considered for high concentration samples such as animal feces, raw sewage and water samples collected immediately following a sewage spill. Second, high concentrations of total DNA (including both target and non-target DNA) can interfere with the droplet partitioning process. It is not advised to use more than 66 ng total DNA without pretreatment by restriction enzymes7. Third, although the EntHF183 duplex dPCR assay is significantly more robust against inhibition than qPCR, this assay can still yield false negative results if severe inhibition prevents PCR amplification from occurring. Inhibition control measures therefore should still be implemented. Standard addition or dilution methods as described previously can be used to assess inhibition7. Last, the duplex assay has only been validated on one dPCR platform, and method validation should be conducted on other digital PCR platforms before use.
This is the first published duplex digital PCR assay that simultaneously quantifies a widely accepted water quality indicator and a popular host-associated marker for fecal contamination7. The demonstrated advantages of the EntHF183 duplex dPCR assay promise other useful applications of digital PCR technology in microbial source tracking, detection of pathogens and antibiotic resistance genes. For example, a general fecal indicator (Enterococcus spp., E. coli, general Bacteroidales) and a host-associated fecal marker (HF183, HumM2, CowM2, LeeSeagull)2 can be paired similarly (as in the EntHF183 duplex dPCR assay) in a duplex dPCR assay for more cost-effective and accurate quantification of general and host-associated fecal contamination. Microbial source tracking markers targeting the same source (or different sources) could also be paired in duplex dPCR assay to increase confidence in detecting the fecal source (or obtain information on multiple fecal sources simultaneously). The higher tolerance against PCR inhibitors could also allow analysis of higher sample volume to increase the probability of detecting pathogens that have low infective doses but are only present in environmental water in low concentrations. Similar methodological advantages of digital PCR over qPCR could also apply to developing digital PCR assays for quantification of antibiotic resistance genes.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors have no acknowledgements.
Materials
| Low Bind Microtubes | Costar | 3207 | For storage of reagents, samples/production of master mixes |
| Nuclease Free Water | FisherSci | BP2484-50 | |
| TE pH 8 buffer | FisherSci | BP2473-100 | |
| Hardshell 96 Well Plate | BioRad | HSP-9601 | For initial master mix and sample inoculation |
| Aluminum Sealing Film | BioRad | 359-0133 | To seal sample plate |
| Droplet Generator | BioRad | 186-3002 | |
| Droplet Generation Oil | BioRad | 186-3005 | |
| Cartridge | BioRad | 186-4008 | |
| DG8 Cartridge Holder | BioRad | 186-3051 | |
| Gasket | BioRad | 186-3009 | |
| 20uL pipet tips | Rainin | GP-L10F | For tansferring sample/master mix to cartidge |
| 200uL pipet tips | Rainin | GP-L200F | For transferring droplets to final Twin.Tec Plate |
| Twin.Tec 96 Well Plate | Eppendorf | 951020320 | For final droplets thermal cycling and reading |
| Pierceable Heat Seal Foil | BioRad | 181-4040 | To seal Twin.Tec plate before thermalcycling |
| PX1 PCR Plate Sealer | BioRad | 181-4000 | Only the thermal cycler is needed, no optics |
| CFX96 Thermalcycler | BioRad | CFX96 and C1000 | |
| QX100 Droplet Reader | BioRad | 186-3001 | |
| Droplet Reader Oil | BioRad | 186-3004 | |
| Droplet PCR Supermix | BioRad | 186-3024 | i.e. the digital PCR mix in manuscript |
| QuantaSoft software (v1.3.2) | Quantasoft | QX100 | For viewing, analyzing, and exporting ddPCR data |
| Entero Forward Primer (Ent F1A) | Operon | GAGAAATTCCAAACGAACTTG | Alternative vendor can be used |
| Entero Reverse Primer (Ent R1) | Operon | CAGTGCTCTACCTCCATCATT | Alternative vendor can be used |
| Entero Probe (GPL813TQ) | Operon | [6-FAM]-TGG TTC TCT CCG AAA TAG CTT TAG GGC TA-[BHQ1] | Alternative vendor can be used, but the florophore has to be FAM |
| HF183 Forward Primer (HF183-1) | Operon | ATCATGAGTTCACATGTCCG | Alternative vendor can be used |
| HF183 Reverse Primer (BthetR1) | Operon | CGTAGGAGTTTGGACCGTGT | Alternative vendor can be used |
| HF183 Probe (BthetP1) | Operon | [6-HEX]-CTGAGAGGAAGGTCCCCC ACATTGGA-[BHQ1] |
Alternative vendor can be used, but the florophore has to be HEX (if using VIC, then the appropriate matric compensation must be chosen. |
| Positive control | Mixture of E.faecalius genomic DNA and HF183 standard plasmid (ordered from IDT). For detailed methods in culturing E. faecalius and sequences of the ordered HF183 plasmid, please see Cao et al 2015 (doi:10.1016/j.watres.2014.12.008). Commercilaly available Enterococcus DNA standards (ATCC 29212Q-FZ) can also be used in the positive control in place of lab-prepared E. faecalis genomic DNA. |
References
- Boehm, A. B., et al. Performance of forty-one microbial source tracking methods: A twenty-seven lab evaluation study. Water Res. 47 (18), 6812-6828 (2013).
- Layton, B. A., et al. Performance of human fecal anaerobe-associated PCR-based assays in a multi-laboratory method evaluation study. Water Res. 47 (18), 6897-6908 (2013).
- Cao, Y., et al. Effect of platform, reference material, and quantification model on enumeration of Enterococcus by quantitative PCR methods. Water Res. 47 (1), 233-241 (2013).
- Sivaganesan, M., Siefring, S., Varma, M., Haugland, R. A. MPN estimation of qPCR target sequence recoveries from whole cell calibrator samples. J. Microbiol. Methods. 87 (3), 343-349 (2011).
- Huggett, J. F., et al. The Digital MIQE Guidelines: Minimum Information for Publication of Quantitative Digital PCR Experiments. Clin. Chem. 59 (6), 892-902 (2013).
- Cao, Y., Raith, M. R., Griffith, J. F. Droplet digital PCR for simultaneous quantification of general and human-associated fecal indicators for water quality assessment. Water Res. 70, 337-349 (2015).
- Sanders, R., Huggett, J. F., Bushell, C. A., Cowen, S., Scott, D. J., Foy, C. A. Evaluation of Digital PCR for Absolute DNA Quantification. Anal. Chem. 83 (17), 6474-6484 (2011).
- Whale, A. S., et al. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acid Res. 40 (11), 82-89 (2012).
- Hindson, C. M., et al. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nature Methods. 10 (10), 1003-1005 (2013).
- Morisset, D., Štebih, D., Milavec, M., Gruden, K., Žel, J. Quantitative analysis of food and feed aamples with droplet digital PCR. PLoS One. 8 (5), e62583 (2013).
- Cao, Y., et al. Evaluation of molecular community analysis methods for discerning fecal sources and human waste. Water Res. 47 (18), 6862-6872 (2013).

