Construction of Cyclic Cell-Penetrating Peptides for Enhanced Penetration of Biological Barriers
Summary
This protocol describes the synthesis of cyclic cell-penetrating peptides with aromatic cross-links and the evaluation of their permeability across biological barriers.
Abstract
Cancer has been a grand challenge in global health. However, the complex tumor microenvironment generally limits the access of therapeutics to deeper tumor cells, leading to tumor recurrence. To conquer the limited penetration of biological barriers, cell-penetrating peptides (CPPs) have been discovered with excellent membrane translocation ability and have emerged as useful molecular transporters for delivering various cargoes into cells. However, conventional linear CPPs generally show compromised proteolytic stability, which limits their permeability across biological barriers. Thus, the development of novel molecular transporters that can penetrate biological barriers and exhibit enhanced proteolytic stability is highly desired to promote drug delivery efficiency in biomedical applications. We have previously synthesized a panel of short cyclic CPPs with aromatic crosslinks, which exhibited superior permeability in cancer cells and tissues compared to their linear counterparts. Here, a concise protocol is described for the synthesis of the fluorescently labeled cyclic polyarginine R8 peptide and its linear counterpart, as well as key steps for investigating their cell permeability.
Introduction
The past few decades have witnessed rapid advances in the development of cell-penetrating peptides (CPPs) for drug delivery. CPPs have been widely used as molecular transporters for the treatment of a range of life-threatening diseases, including neurological disorders1,2, heart diseases3, diabetes4, dermatosis5, and cancer6,7. Cancer remains a global health burden accompanied by a high rate of morbidity and mortality despite widespread research efforts8. A serious obstacle to treating cancer is the limited access of therapeutics to deeper tumor cells due to physiological barriers such as compact extracellular matrix (ECM), abnormal tumor vasculature, multiple membrane barriers, and high interstitial fluid pressure (IFP)9. Thus, developing novel CPPs with superior ability to deliver cargoes across biological barriers is considered an essential strategy for cancer treatment10,11.
CPPs can be categorized into cationic, amphipathic, and hydrophobic CPPs in terms of their physicochemical properties12. Among these, the positively charged HIV-TAT peptide and the synthetic polyarginine are of considerable importance in biomedical research and have been extensively studied to facilitate intracellular drug delivery13. Tunnemann et al. reported that a minimum length of eight arginines is essential for efficient cell penetration of the synthetic polyarginine peptides, based on a cell permeability study conducted using R3 to R12 peptides14. However, these CPPs generally have short plasma half-lives due to their rapid hydrolysis in vivo. In addition, little is known regarding the optimization of the chemical structure of CPPs to increase their trans-barrier ability as it is challenging to penetrate multiple cell membranes15. Thus, the development of novel molecular transporters capable of penetrating biological barriers is strongly desired to enhance drug delivery efficiency. In 2020, Komin et al.16 discovered a CPP called CL peptide, which contains a helix motif (RLLRLLR) and a polyarginine tail (R7) for crossing the epithelial monolayer. A set of CL peptide variants were also synthesized by altering the helical pattern. This exploration could be a significant guide for the development of novel CPPs for the delivery of cargoes across biological barriers. Moreover, Dietrich et al. optimized the cell permeability of the StAX peptide, inhibiting the Wnt/β-catenin signaling pathway by increasing the overall hydrophobicity of the peptides17.
Conformational restriction of unstructured linear peptides by cyclization is an effective way to enhance their proteolytic stability and permeablity18,19,20. The structural reinforcement increases the protease resistance of cyclic peptides, making them more stable in vivo compared to their linear counterparts. In addition, the cyclization of peptides can potentially mask the polar peptide backbone by promoting intramolecular hydrogen bonding, thus increasing the membrane permeability of the peptides21. In the past two decades, chemoselective cyclization methods have become effective strategies for the construction of cyclic peptides with different architectures, such as all-hydrocarbon, lactam, triazole, m-xylene, perfluoroaryl, and other cross-links22,23. The biological barrier imposed by the sophisticated tumor microenvironment could reduce the penetration of drugs in solid tumors24. We have previously found that the cyclic CPPs displayed superior resistance to enzymatic digestion over their linear counterparts20. Furthermore, the overall hydrophobicity of the peptides is critical for their enhanced cell permeability22. Based on the studies discussed above, the combination of a positively charged pattern, elevated overall hydrophobicity, and enhanced proteolysis stability can be hypothesized to increase the permeability of CPPs across biological barriers. In a recent study, we identified two cyclic CPPs with aromatic crosslinks at positions i and i+7 that exhibit improved permeability in tumor cells and tissues compared to their linear counterparts15. Here, a concise synthetic protocol for the synthesis of fluorescently labeled cyclic CPPs and the key steps to investigate their permeability are presented.
Protocol
1. Equipment preparation
NOTE: Carry out all the procedures in an operating fume hood with suitable personal protective equipment.
- Assemble the manual peptide synthesis apparatus in the fume hood (Figure 1). Place the three-way stopcocks (see Table of Materials) onto the vacuum manifold (see Table of Materials) and connect to the nitrogen (N2). Make sure to cap the unused inlets.
- Attach a 10 mL polypropylene column (see Table of Materials) onto the three-way stopcocks. Drain the reaction mixture or solvents from the polypropylene column using a rubber pipette bulb or a vacuum via a waste trap.
2. Synthesis of FITC-labeled linear R8 peptide (FITC-R8) and FITC-labeled stapled R8 peptide (FITC-sR8-4)
NOTE: The peptides were synthesized according to a standard Fmoc-based solid-phase peptide synthesis (SPPS) protocol25. The 4-(2',4'-Dimethoxypheyl-Fmoc-aminomethyl)-phenoxyacetamido-norleucyl-MBHA resin (rink amide MBHA resin, see Table of Materials) was used throughout the study.
CAUTION: N, N-dimethylformamide (DMF), N, N-diisopropylethylamine (DIPEA), morpholine, and dichloromethane (DCM) are all colorless and are damaging if inhaled or absorbed through the skin. Ether is extremely flammable. 1,2-Ethanedithiol (EDT) is a particularly odorous substance. Trifluoroacetic acid (TFA) is highly corrosive, and its acidity is 105 times that of acetic acid. Consequently, all reagents and chemicals are supposed to be dealt with using protective equipment in a fume hood.
- Prepare the resin for peptide synthesis.
- Calculate the mass of resin needed for the synthesis:Mass of resin (mg) = scale (mmol) / resin loading capacity (mmol/g) × 1,000 (mg/g)
NOTE: For example, the mass of rink amide MBHA resin (0.572 mmol/g) for 0.2 mmol = 0.2 mmol / 0.572 mmol/g × 1,000 mg/g = 350 mg. - Add 4-5 mL of DMF to the required amount of resin and transfer into the 10 mL polypropylene column (step 1.2) with gentle N2 bubbling for 30 min to swell the resin adequately, and then drain the DMF.
- Add 4-5 mL of 50% morpholine/DMF (v/v) to the resin, gently bubble N2 for 30 min 2x to remove the N-terminal Fmoc group, and then drain the mixture. Afterward, wash the resin thoroughly 3x by adding 4-5 mL of DMF to the column and bubbling with N2 for at least 1 min each time. Continue to wash the resin with DCM (3x) and DMF (3x) in the same way.
- Calculate the mass of resin needed for the synthesis:Mass of resin (mg) = scale (mmol) / resin loading capacity (mmol/g) × 1,000 (mg/g)
- Perform Fmoc-protected amino acid coupling as described below.
NOTE: The coupling of arginine in a 0.2 mmol scale manual synthesis is described here as an example.- Dissolve Fmoc-Arg (Pbf)-OH (5 equiv., 648.8 mg) and 2-(7-Azobenzotriazole)-N, N, N', N'-tetramethyluronium hexafluorophosphate (HATU, 4.9 equiv., 372.6 mg) in 5 mL of DMF in a centrifuge tube.
- Add DIPEA (10 equiv., 348.4 µL) to activate the coupling reaction and then transfer the reaction mixture to the 10 mL polypropylene column with resin (prepared in step 2.1.3). Then, gently agitate the mixture with N2 bubbling for 1-2 h.
- Repeat the coupling reaction (step 2.2.1 and step 2.2.2) once.
- After completion of the coupling, drain the reaction mixture and wash the resin sequentially with DMF, DCM, and DMF 3x each for at least 1 min each time.
- Perform coupling of each amino acid in ordered steps: Add 4-5 mL of 50% morpholine/DMF (v/v) to the resin, gently bubble with N2 for 30 min 2x to remove the Nα-Fmoc group, then wash the resin (as shown in step 2.2.4), and proceed to couple the next amino acid (as shown in step 2.2.1 and step 2.2.2). Proceed with several cycles of this step to achieve synthesis of the desired peptide.
NOTE: This process can be paused here. Condense the resin with methanol and dry the resin with a continuous flow of N2. Cap the polypropylene column and then store the resin at 4 °C for a few days (or at −20°C for longer storage). Swell the resin with 4-5 mL of DMF for 0.5-1 h before starting a new synthesis. If directly proceeding to the next step, there is no need to condense the resin.
- Label the peptides with fluorescein isothiocyanate (FITC) as described below.
- Couple the beta-alanine as a spacer for FITC labeling using the same process as used for amino acid coupling in step 2.2.
- Perform FITC labeling of peptides on the resin by adding a mixture of FITC (5 equiv.), DIPEA (10 equiv.), and DMF to the polypropylene column and reacting in the dark for 8 h.
- For the synthesis of FITC-sR8-4, perform cyclization of the linear peptide as described below.
- Add a mixture of TFA/triisopropylsilane (TIS)/DCM (3/5/92, v/v/v) to the polypropylene column for 2 min to selectively remove the Cys (Trt) protecting group, and then drain the mixture. Repeat the above procedure until the yellowish solution becomes colorless to completely remove the Trt protecting group.
- Subsequently, perform sequential washes of the resin with DMF and DCM at least 3x. Afterward, dissolve 4,4'-bis(bromomethyl)biphenyl (2 equiv.) in DMF with DIPEA (4 equiv.), add it to the column, and react for 4 h.
- Cleave the peptides as described: After the completion of peptide synthesis, wash the resin with 4-5 mL of methanol twice for 5 min each and dry it with a continuous flow of N2. Treat the resin with an effective cleavage cocktail TFA/TIS/H2O (95/2.5/2.5, v/v/v), or TFA/TIS/EDT/H2O (92.5/2.5/2.5/2.5, v/v/v/v) for peptides containing cysteines, using approximately 1 mL of cleavage cocktail for per 100 mg of resin. Treat the peptide-bound resin for 2-3 h to cleave the peptide and then remove the TFA carefully with a stream of N2.
- To obtain the crude peptides, add 4-5 mL of diethyl ether to the cleaved peptide preparation to precipitate the crude peptides and centrifuge at 10,000 × g for 4 min. Carefully discard the supernatant and air-dry the peptide for 3 min in an effective fume hood.
- Analysis of the peptides: Dissolve a small-scale crude peptide (cleaved from approximately 10 mg of peptide-bound resin) in 800 µL of acetonitrile (ACN)/H2O (1/1, v/v) and then analyze using reverse-phase high-performance liquid chromatography (RP-HPLC) and liquid chromatography-mass spectrometry (LC-MS) (see Table of Materials).
- Purify the peptides using RP-HPLC and LC-MS.
- Dissolve 50 mg of crude peptide product in 4 mL of ACN/H2O (1/1, v/v), and inject the solution into an RP-HPLC system equipped with a C18 column (4.6 mm x 150 mm, pore size: 120 Å, particle size: 4 µm; see Table of Materials). Elute the peptide using a mobile phase containing 0.1% TFA/H2O (v/v) and ACN, with a gradient of 10% to 90% ACN over 30 min. Conventional peptides were detected at 220 nm and FITC-labeled peptides at 494 nm.
- Collect the fractions corresponding to the major peptide peak as identified by MS, and then lyophilize the desired peptide fractions. Store the purified peptide at −20 °C.
NOTE: The found m/z of the purified FITC-R8 are as follows: [M + 3 H]3+: 576.63; [M + 4 H]4+: 432.72; [M + 5 H]5+: 346.39; [M + 6 H]6+: 288.85. The found m/z of the purified FITC-sR8-4 are as follows: [M + 3 H]3+: 704.74; [M + 4 H]4+: 528.77; [M + 5 H]5+: 423.34; [M + 6 H]6+: 352.91. MS analytical conditions: instrument: ESI (probe bias: +4.5 kV; detector: 1.2 kV); nebulizer gas flow: 1.5 L/min; curved desolvation line (CDL): −20 V; CDL temperature: 250 °C; block temperature: 400 °C; flow rate: 0.2 mL/min; mobile phase: 50% H2O/50% ACN.
3. Quantification of the FITC-labeled peptides
- Dissolve a small amount of purified peptide in DMSO as stock solution (e.g., 40 µmol/mL).
- Measure the absorbance at 494 nm (A494) of 2 µL of the stock solution in 498 µL of 10 mM phosphate buffered saline (1x PBS, pH 7.4) with a microtiter plate (see Table of Materials) using a multi-technology microplate reader (see Table of Materials). The dilution factor is 500 µL/ 2 µL = 250, and the path length of the microtiter plate is 0.5 mm.
- Calculate the concentration of the stock solution using the following formula:
Concentration (mM) = A494 × dilution factor / 0.05 (cm) / 77,000 (cm−1·M−1) × 1,000 (mM·M−1) - Adjust to a proper dilution so that the measured A494 value is between 0.1 and 1.0.
NOTE: The measurements should be repeated several times to ensure that the measured concentration is accurate. The extinction coefficient of 77,000 cm−1·M−1 arises from the FITC group.
4. Stability of peptides in fetal bovine serum (FBS)
- Incubate the peptide at a concentration of 100 µM with 250 µL of 25% FBS/H2O (v/v) at 37 °C. After incubating for 0 h, 1 h, 2 h, and 4 h, take 10 µL aliquots and then add 150 µl of 12% trichloroacetic acid dissolved in H2O/ACN (1/3, v/v) to precipitate the serum proteins.
- Centrifuge the samples at 10,000 × g for 5 min and analyze the supernatant using HPLC (as described in step 2.8) to determine the extent of peptide degradation.
- Calculate the ratio of peak area at 1 h, 2 h, and 4 h to that at 0 h to obtain the fraction of undegraded peptide at the corresponding time. The result is the average of three parallel samples.
5. Cellular uptake of the peptides
- Fluorescence microscopic imaging
- Place a round coverslip into a 12-well plate. Then, inoculate 1 x 105 cells uniformly on coverslips and culture overnight with 2 mL of medium. Remove the medium and wash the cells 3x with 1 mL of PBS.
NOTE: In this study, HeLa cells and 4T1 cells were cultured in Dulbecco's Modified Eagle Medium (DMEM, see Table of Materials) supplemented with 10% FBS in a 37 °C humidified incubator containing 5% CO2. - Incubate the cells with 1 mL of 3 µM FITC-labeled peptides in FBS-free DMEM for 1 h at 37 °C. Afterward, remove the peptide-containing medium and wash the cells 3x with 1 mL of PBS.
- Stain the cells with 1 mL of Hoechst 33258 for 15 min. Observe the internalization of each peptide using a fluorescence microscope (see Table of Materials) with the same fluorescence intensity and exposure time.
NOTE: The following settings were used for fluorescence microscopy. Objective: Plan-Apochromat: 63 x/1.40 Oil DIC M27; Channel 1 for FITC: excitation filter: 450-490 nm, emission filter: 500-550 nm, exposure time: 230 ms; Channel 2 for Hoechst 33258: excitation filter: 335-383 nm, emission filter: 420-470 nm, exposure time: 27 ms.
- Place a round coverslip into a 12-well plate. Then, inoculate 1 x 105 cells uniformly on coverslips and culture overnight with 2 mL of medium. Remove the medium and wash the cells 3x with 1 mL of PBS.
- Flow cytometry analysis
- Inoculate 5 x 105 HeLa cells uniformly in 12-well plates and culture in DMEM for 24 h at 37 °C. Afterward, remove the medium and wash the cells 3x with 1 mL of PBS.
- Incubate the cells with 1 mL of 3 µM FITC-labeled peptides in FBS-free DMEM for 1 h at 37 °C. Remove the peptide-containing medium, dissociate the cells with 0.25% (w/v) trypsin and 0.53 mM EDTA in PBS for 5 min, and then collect the cells by centrifugation at 306 × g for 4 min. Wash the cell pellet with PBS.
- Incubate the cells with 1 mL of 0.05% (w/v) trypan blue in PBS for 3 min to quench the surface-bound fluorescence, and perform quantitative analysis of intracellular fluorescence using a flow cytometer (see Table of Materials).
NOTE: Flow cytometry settings: excitation: 488 nm, emission: 530 nm. Trypan blue can also quench the fluorescence of dead cells and help distinguish live/dead cells during analysis of peptide uptake. - Treat and assay 4T1 cells using flow cytometry following the same protocol as described for HeLa cells. Collect 5 x 105 cells per sample and set up three parallel samples per group.
6. Exploration of the cell-to-cell penetration of the peptides using transwell models
- Inoculate 1 x 105 HeLa cells in 2 mL of DMEM in a 12-well chamber with a tissue culture plate insert (see Table of Materials) and incubate for 24 h in a 37 °C humidified incubator containing 5% CO2. Afterward, remove the medium and incubate the cells in the chambers with 1 mL of 10 µM FITC-R8 or FITC-sR8-4 (purified using HPLC) in FBS-free DMEM for 1 h.
- Remove the medium containing the peptides and wash the cells 3x with 1 mL of PBS. Add 1 mL of fresh FBS-free DMEM to the chambers and then co-incubate the HeLa cells in the chamber with the tissue culture plate insert with the HeLa cells on the round coverslips at the bottom for 2 h.
- Fix the HeLa cells on the round coverslips with 2.5% glutaraldehyde for 15 min and then stain the cells with DAPI for 15 min. Then, observe the HeLa cells on the coverslips under a fluorescence microscope. Treat and assay 4T1 cells using the same protocol as that used for HeLa cells.
Representative Results
In this protocol, a synthetic procedure to constrain the linear polyarginine R8 into its cyclic form was presented. The SPPS was conducted manually using a simple apparatus (Figure 1). The detailed synthetic process of SPPS is shown in Figure 2. Briefly, the resin was sufficiently swelled, followed by deprotection of the Nα-Fmoc protecting group. Then, the Nα-Fmoc-protected amino acid was anchored on the resin until the completion of the peptide assembly (steps 1-4 in Figure 2). Then, the crude peptides were cleaved from the resin by the cleavage cocktail (step 5 in Figure 2). FITC was used to label the peptides to synthesize fluorescently labeled cyclic CPPs and track their permeability across biological barriers. Subsequently, the trityl protecting groups of cysteines were selectively deprotected on the resin, followed by peptide cyclization with 4,4'-bis(bromomethyl)biphenyl cross-link (Figure 3A). The HPLC and MS spectra of FITC-R8 and FITC-sR8-4 are shown in Figure 3B. The retention time of FITC-sR8-4 was substantially longer than that of the linear analog, indicating enhanced overall hydrophobicity of the peptide after cyclization with the hydrophobic cross-link. Furthermore, as shown in Figure 3C, the cyclic R8 remained 77.3% intact after incubation with 25% FBS for 4 h, while its linear counterpart was mostly degraded, suggesting enhanced proteolytic stability of the cyclic R8 peptide. In the subsequent cell-based studies, cells treated with cyclic R8 with aromatic crosslink exhibited higher intracellular fluorescence than those treated with its linear counterpart, as demonstrated by live-cell fluorescence microscopy imaging (Figure 4A). Similar results were obtained with flow cytometry analysis (Figure 4B). To further investigate whether cyclic R8 confers enhanced cell-to-cell penetration, transwell models were used to simulate the barrier permeability of the peptides from one cell layer to another. The cyclic R8 clearly exhibited higher trans-barrier penetration than the linear R8 peptide, as indicated by a significant increase in the intracellular fluorescence (Figure 4C). To sum up, the cyclic R8 peptide exhibited superior permeability across biological barriers over its linear counterpart.

Figure 1: Equipment setup for the manual peptide synthesis apparatus. A 10 mL polypropylene column is set up on the vacuum manifold using a three-way stop valve. N2 is used for agitation. Please click here to view a larger version of this figure.
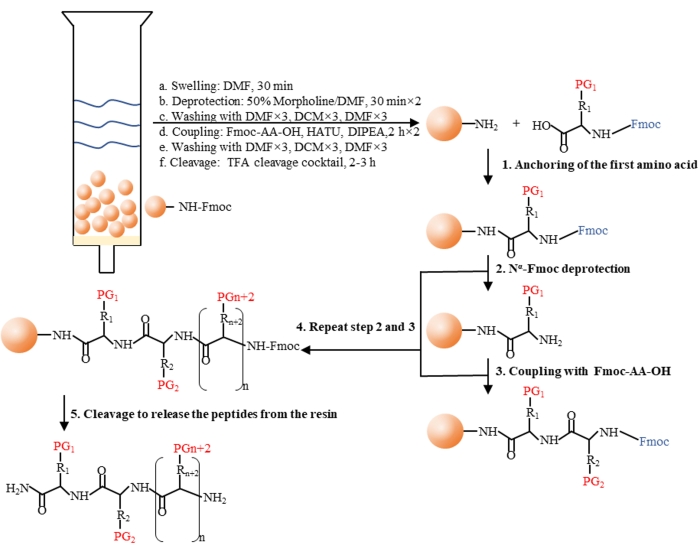
Figure 2: General procedure of Fmoc solid-phase peptide synthesis (SPPS). An Nα-Fmoc-protected amino acid is anchored to the 4-(2',4'-Dimethoxypheyl-Fmoc-aminomethyl)-phenoxyacetamido-norleucyl-MBHA resin (rink amide MBHA resin) (step 1), followed by deprotection of the Nα-Fmoc protecting groups of the amino acids (step 2) and subsequent amino acid coupling (step 3). Step 2 and step 3 are repeated several times to synthesize the desired peptide (step 4). After the completion of synthesis, a cleavage cocktail is added to remove the side chain protecting groups and cleave the desired peptide from the resin (step 5). Abbreviations: DMF = N, N-dimethylformamide; DCM = dichloromethane; HATU = 2-(7-Azobenzotriazole)-N, N, N', N'-tetramethyluronium hexafluorophosphate; DIPEA = N, N-diisopropylethylamine; TFA = trifluoroacetic acid. Please click here to view a larger version of this figure.
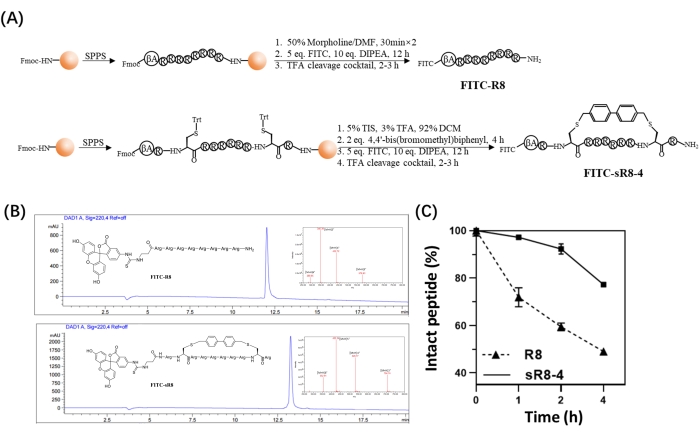
Figure 3: Synthesis of FITC-labeled linear R8 peptide (FITC-R8) and FITC-labeled stapled R8 peptide (FITC-sR8-4) using solid-phase peptide synthesis (SPPS). (A) Schematic diagram of the synthesis of FITC-R8 and FITC-sR8-4. (B) HPLC and MS spectra (inset) of FITC-R8 and FITC-sR8-4. (C) Stability of FITC-R8 and FITC-sR8-4 in the presence of 25% FBS. Intact peptide (%) refers to the fraction of undegraded peptide. This figure has been modified from Shi et al.15. Please click here to view a larger version of this figure.
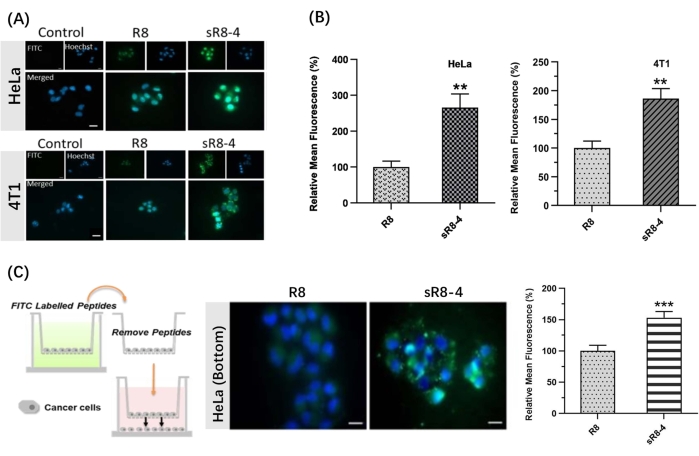
Figure 4: Penetration of FITC-labeled linear R8 peptide (FITC-R8) and FITC-labeled stapled R8 peptide (FITC-sR8-4). (A) Live-cell fluorescence microscopy images of HeLa cells and 4T1 cells after 1 h incubation with 3 µM FITC-R8 and FITC-sR8-4. FITC (green), Hoechst (blue). Scale bar = 20 µm. (B) Relative mean fluorescence (with respect to the linear R8 peptide), mean ± s.d., and n = 3; (C) Cell-to-cell penetration of FITC-R8 and FITC-sR8-4 in a transwell model using HeLa cells. Live-cell fluorescence microscopy images (scale bar = 20 µm), and relative mean fluorescence (with respect to the linear R8 peptide), mean ± s.d., and n = 3. ** P < 0.01, *** P < 0.001. This figure has been modified from Shi et al.15. Please click here to view a larger version of this figure.
Discussion
The chemical stabilization of peptides by incorporating conformational constraints has proven to be an effective strategy for improving the stability and cell permeability of the peptide26. In this protocol, a step-by-step procedure is described for the synthesis of cyclic CPPs with aromatic cross-links and the evaluation of their permeability across biological barriers. Compared to the hydrophilic lactam or triazole cross-links22,27, the incorporation of aromatic cross-links (used in this study) improves the overall hydrophobicity of the CPPs, thereby significantly increasing their cell permeability. On the other hand, peptide cyclization can be easily achieved through substitution reactions with cysteines without requiring any metal catalysts. In this protocol, the cyclization of the CPPs was conducted on resin; however, the cyclization efficiency also depends on the specific sequences and lengths of the peptides due to steric effects, which may result in the formation of a dimeric byproduct28. In such a case, using a resin with a lower loading capacity would be helpful. In addition, it is also recommended to cyclize these specific peptides under dilute concentrations in the solution phase29.
There are a few critical points in this protocol. First, the cleavage cocktail TFA/TIS/EDT/H2O (92.5/2.5/2.5/2.5, v/v/v/v) is used for the cleavage of cysteine-containing peptides to prevent oxidation of the sulfhydryl group. Second, it is suggested to perform a small-scale preliminary study to obtain the appropriate cleavage condition. The optimal time required to cleave the peptides from the resin is 2-3 h, with a longer cleavage time (more than 5 h) tending to produce more unidentified byproducts. The peptide synthesis could be monitored by LC-MS to optimize the cleavage time. Third, FITC labeling should be done in the dark to avoid fluorescence quenching.
Furthermore, trypan blue should be used to quench the surface-bound fluorescence as flow cytometry analysis cannot distinguish intracellular or surface-bound fluorescence. This will help to specifically quantify the peptide internalized by the cancer cells27. In addition, as cationic peptides may also cause non-specific membrane lysis30, hemolytic activity and cell viability could also be conducted to evaluate the toxicity of the cyclic CPPs.
Cyclic CPPs constitute one of the effective drug delivery vehicles for conquering biological barriers. However, the membrane interaction and perturbation of cationic CPPs generally lead to potential non-specific cytotoxicity31. Further efforts will be devoted to understanding the detailed penetration mechanism, which should aid the discovery of the next generation of cyclic CPPs to penetrate biological barriers with minimal cytotoxicity. These highly active and stable CPPs hold great promise for improving the treatment of important life-threatening diseases.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work is supported by the Natural Science Foundation of China (21708031), China Postdoctoral Science Foundation (BX20180264, 2018M643519), and the Fundamental Research Funds for the Central Universities (2682021ZTPY075).
Materials
| 1,2-ethanedithiol | Aladdin | K1722093 | stench |
| 2-(7-Azobenzotriazole)-N,N,N',N'-tetramethyluronium hexafluorophosphate (HATU) | HEOWNS | A-0443697 | |
| 4,4'-bis(bromomethyl)biphenyl | TCI | B1921 | |
| 4T1 cells | ATCC | 4T1 cells were cultured in DMEM medium supplemented with 10% FBS (Hyclone) in a 37 °C humidified incubator containing 5% CO2. | |
| Acetonitrile | Adamas | 1484971 | toxicity |
| Dichloromethane | Energy | W330229 | skin harmful |
| Diethyl ether | Aldrich | 673811 | flammable |
| Dimethyl sulfoxide | Beyotime | ST038 | skin harmful |
| Dulbecco’s Modified Eagle Medium (DMEM) | Gibco | ||
| Electrospray Ionization Mass Spectrometer | Waters | G2-S Tof | |
| Ethylene Diamine Tetraacetic Acid (EDTA) | BioFroxx | 1340 | |
| Fetal bovine serum (FBS) | HyClone | ||
| Flow cytometer | Beckman Coulter | CytoFLEX | |
| Fluorescein isothiocyanate isomer (FITC) | Energy | E0801812500 | |
| Fluorescent microscope | Carl Zeiss | Axio Observer 7 | |
| Fmoc-Arg(Pbf)-OH | HEOWNS | F-81070 | |
| Fmoc-Cys(Trt)-OH | GL Biochem | GLS201115-35202 | |
| Fmoc-βAla-OH | Adamas | 51341C | |
| HeLa cells | ATCC | HeLa cells were cultured in DMEM supplemented with 10% FBS (Hyclone) in a 37 °C humidified incubator containing 5% CO2. | |
| High-Performance Liquid Chromatography | Agilent | Agilent 1260 | |
| High-Performance Liquid Chromatography column | Agilent | Poroshell EC-C18 120, 4.6 × 150 mm (pore size 120 Å, particle size 4 μm) | |
| Lyophilizer | SP Scientific | Vir Tis | |
| Methanol | Aldrich | 9758 | toxicity |
| Microtiter plate | Thermo μdrop plate | N12391 | |
| Morpholine | HEOWNS | M99040 | irritant |
| Multi-technology microplate reader | Thermo | VARIOSKAN LUX | |
| N,N-Diisopropylethylamine | HEOWNS | E-81416 | irritant |
| N,N-Dimethyl formamide | Energy | B020051 | harmful to skin |
| Poly-Prep column | Bio-Rad | 7321010 | polypropylene chromatography columns |
| Rink Amide MBHA resin (0.572 mmol/g) | GL Biochem | GLS180301-49101 | |
| Three-way stopcocks | Bio-Rad | 7328107 | |
| Tissue culture plate insert | LABSELECT | 14211 | |
| Trifluoroacetic acid | HEOWNS | T63278 | corrosive |
| Triisopropylsilane | HEOWNS | T-0284475 | |
| Trypsin | BioFroxx | 1004 | |
| Vacuum manifold | Promega | A7231 |
References
- Zhang, L., et al. Brain-targeted dual site-selective functionalized poly(β-amino esters) delivery platform for nerve regeneration. Nano Letters. 21 (7), 3007-3015 (2021).
- Park, T. E., et al. Enhanced BBB permeability of osmotically active poly(mannitol-co-PEI) modified with rabies virus glycoprotein via selective stimulation of caveolar endocytosis for RNAi therapeutics in Alzheimer’s disease. Biomaterials. 38, 61-71 (2015).
- Bian, J., et al. Effect of cell-based intercellular delivery of transcription factor GATA4 on ischemic cardiomyopathy. Circulation Research. 100 (11), 1626-1633 (2007).
- He, H., et al. The use of low molecular weight protamine chemical chimera to enhance monomeric insulin intestinal absorption. Biomaterials. 34 (31), 7733-7743 (2013).
- Kim, D., et al. A specific STAT3-binding peptide exerts antiproliferative effects and antitumor activity by inhibiting STAT3 phosphorylation and signaling. 癌症研究. 74 (8), 2144-2151 (2014).
- Yang, Y., et al. PEGylated liposomes with NGR ligand and heat-activable cell-penetrating peptide-doxorubicin conjugate for tumor-specific therapy. Biomaterials. 35 (14), 4368-4381 (2014).
- Wei, Y., et al. Intracellular paclitaxel delivery facilitated by a dual-functional CPP with a hydrophobic hairpin tail. ACS Applied Materials and Interfaces. 13 (4), 4853-4860 (2021).
- Vasan, N., Baselga, J., Hyman, D. M. A view on drug resistance in cancer. Nature. 575 (7782), 299-309 (2019).
- Cong, Y., et al. Microenvironment-induced in situ self-assembly of polymer-peptide conjugates that attack solid tumors deeply. Angewandte Chemie International Edition. 131 (14), 4680-4685 (2019).
- Blanco, E., Shen, H., Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nature Biotechnology. 33 (9), 941-951 (2015).
- Tian, Y., Zhou, S. Advances in cell-penetrating peptides and their functionalization of polymeric nanoplatforms for drug delivery. Wiley Interdisciplinary Reviews. Nanomedicine and Nanobiotechnology. 13 (2), 1-12 (2021).
- Milletti, F. Cell-penetrating peptides: Classes, origin, and current landscape. Drug Discovery Today. 17 (15-16), 850-860 (2012).
- Turner, J. J., et al. Cell-penetrating peptide conjugates of peptide nucleic acids (PNA) as inhibitors of HIV-1 Tat-dependent trans-activation in cells. Nucleic Acids Research. 33 (21), 6837-6849 (2005).
- Tunnemann, G., et al. Live-cell analysis of cell penetration ability and toxicity of oligo-arginines. Journal of Peptide Science. 14 (4), 469-476 (2008).
- Shi, M., et al. Stapling of short cell-penetrating peptides for enhanced tumor cell-and-tissue dual-penetration. Chemical Communications. 58 (14), 2299-2302 (2022).
- Komin, A., et al. A peptide for transcellular cargo delivery: Structure-function relationship and mechanism of action. Journal of Controlled Release. 324, 633-643 (2020).
- Dietrich, L., et al. Cell permeable stapled peptide inhibitor of Wnt signaling that targets β-catenin protein-protein interactions. Cell Chemical Biology. 24 (8), 958-968 (2017).
- Tian, Y., et al. Stapling of unprotected helical peptides via photo-induced intramolecular thiol-yne hydrothiolation. Chemical Science. 7 (5), 3325-3330 (2016).
- De Araujo, A. D., et al. Comparative α-helicity of cyclic pentapeptides in water. Angewandte Chemie International Edition. 53 (27), 6965-6969 (2014).
- Chu, Q., et al. Towards understanding cell penetration by stapled peptides. Medicinal Chemistry Communications. 6 (1), 111-119 (2015).
- Bock, J. E., Gavenonis, J., Kritzer, J. A. Getting in shape: Controlling peptide bioactivity and bioavailability using conformational constraints. ACS Chemical Biology. 8 (3), 488-499 (2013).
- Tian, Y., et al. Effect of stapling architecture on physiochemical properties and cell permeability of stapled α-helical peptides: A comparative study. ChemBioChem. 18 (21), 2087-2093 (2017).
- White, C. J., Yudin, A. K. Contemporary strategies for peptide macrocyclization. Nature Chemistry. 3 (7), 509-524 (2011).
- Jain, R. K. Normalization of tumor vasculature: An emerging concept in antiangiogenic therapy. Science. 307 (5706), 58-62 (2005).
- Patgiri, A., Menzenski, M. Z., Mahon, A. B., Arora, P. S. Solid-phase synthesis of short α-helices stabilized by the hydrogen bond surrogate approach. Nature Protocols. 5 (11), 1857-1865 (2010).
- Baek, S., et al. Structure of the stapled p53 peptide bound to Mdm2. Journal of the American Chemical Society. 134 (1), 103-106 (2012).
- Traboulsi, H., et al. Macrocyclic cell penetrating peptides: A study of structure-penetration properties. Bioconjugate Chemistry. 26 (3), 405-411 (2015).
- Tian, Y., et al. A proline-derived transannular N-cap for nucleation of short α-helical peptides. Chemical Communications. 52 (59), 9275-9278 (2016).
- Muppidi, A., et al. Rational design of proteolytically stable, cell-permeable peptide-based selective Mcl-1 inhibitors. Journal of the American Chemical Society. 134 (36), 14734-14737 (2012).
- Wiradharma, N., et al. Synthetic cationic amphiphilic α-helical peptides as antimicrobial agents. Biomaterials. 32 (8), 2204-2212 (2011).
- Jones, A. T., Sayers, E. J. Cell entry of cell penetrating peptides: Tales of tails wagging dogs. Journal of Controlled Release. 161 (2), 582-591 (2012).

