Confocal Imaging of Double-Stranded RNA and Pattern Recognition Receptors in Negative-Sense RNA Virus Infection
Summary
Double-stranded RNA produced during RNA virus replication can be recognized by pattern recognition receptors to induce an innate immune response. For negative-sense RNA viruses, the interaction between the low-level dsRNA and PRRs remains unclear. We have developed a confocal microscopy method to visualize arenavirus dsRNA and PRR in individual cells.
Abstract
Double-stranded (ds) RNA is produced as a replicative intermediate during RNA virus infection. Recognition of dsRNA by host pattern recognition receptors (PRRs) such as the retinoic acid (RIG-I) like receptors (RLRs) RIG-I and melanoma differentiation-associated protein 5 (MDA-5) leads to the induction of the innate immune response. The formation and intracellular distribution of dsRNA in positive-sense RNA virus infection has been well characterized by microscopy. Many negative-sense RNA viruses, including some arenaviruses, trigger the innate immune response during infection. However, negative-sense RNA viruses were thought to produce low levels of dsRNA, which hinders the imaging study of PRR recognition of viral dsRNA. Additionally, infection experiments with highly pathogenic arenaviruses must be performed in high containment biosafety level facilities (BSL-4). The interaction between viral RNA and PRRs for highly pathogenic RNA virus is largely unknown due to the additional technical challenges that researchers need to face in the BSL-4 facilities. Recently, a monoclonal antibody (Mab) (clone 9D5) originally used for pan-enterovirus detection has been found to specifically detect dsRNA with a higher sensitivity than the traditional J2 or K1 anti-dsRNA antibodies. Herein, by utilizing the 9D5 antibody, we describe a confocal microscopy protocol that has been used successfully to visualize dsRNA, viral protein and PRR simultaneously in individual cells infected by arenavirus. The protocol is also suitable for imaging studies of dsRNA and PRR distribution in pathogenic arenavirus infected cells in BSL4 facilities.
Introduction
The initial step of the induction of the innate immune response is host recognition of double-stranded (ds) RNA by the pattern recognition receptors (PRRs) such as the retinoic acid (RIG-I) like receptors (RLRs) RIG-I and melanoma differentiation-associated protein 5 (MDA-5)1. For positive-sense RNA viruses, dsRNA can usually be readily detected using the J2 or K1 anti-dsRNA monoclonal antibodies (Mab)2. Interaction between dsRNA and PRRs in positive-strand RNA viruses, such as picornavirus, has been characterized using confocal microscopy3. However, for negative-sense RNA virus, visualization and characterization of PRR and dsRNA interaction has been hindered by the lack of sensitive antibodies to dsRNA. RNA fluorescent in situ hybridization (FISH) has been applied to the visualization of viral RNA and PRRs4. Nevertheless, the FISH methodology requires the knowledge of the target RNA sequence and may not be compatible with PRR co-staining. Recently, the 9D5 Mab, which was originally developed for the diagnosis of pan-enterovirus infection, was found to be more sensitive than the J2 Mab and can readily detect dsRNA in negative-sense RNA virus infection5,6. Thus, Mab 9D5 is a novel and useful tool to study viral replication and the interaction between PRR and viral RNA for negative-sense RNA virus.
Arenaviruses are a family of single-stranded, negative-sense RNA viruses, which include several human pathogens, such as Lassa virus (LASV), Junín virus (JUNV) and Machupo virus (MACV), that cause severe hemorrhagic fever diseases in humans7. Clinical data from severe and fatal cases of Argentine hemorrhagic fever caused by the New World arenavirus JUNV exhibit unusually high levels of serum IFN-α8,9. We have shown that the pathogenic NW arenaviruses (JUNV and MACV), but not the pathogenic Old World arenavirus, LASV, induce a type I interferon (IFN) response in human monocyte-derived dendritic cells10. Furthermore, RIG-I is one of the sensors mediating type I IFN response in JUNV-infected cells11. We also found that the protein kinase R (PKR) receptor, which is traditionally known for dsRNA recognition, is activated in pathogenic NW arenavirus infection12. To further understand the mechanism of virus-specific IFN response during arenavirus infection, we aimed to develop a protocol to visualize the interaction between viral dsRNA and the cytoplasmic PRRs.
Infection experiments with pathogenic JUNV, MACV and LASV have to be performed in biosafety level 4 (BSL-4) facilities. Thus, in addition to the presumably low level of dsRNA formed in arenavirus infection, meeting the biosafety requirements is another technique challenge when performing imaging studies for these highly pathogenic viruses. By utilizing the 9D5 antibody and the Candid1# vaccine strain of JUNV, a confocal microscopy-based protocol is described in this report, which has been used successfully to visualize dsRNA, viral protein and PRR simultaneously in cells infected by arenavirus in BSL2 labs. The protocol is also suitable for visualization of intracellular distribution of dsRNA and PRR during pathogenic arenavirus infection in BSL4 facilities.
Protocol
1. Preparation of A549 cells and JUNV infection
- Seed 2 x 105 human lung epithelial A549 cells onto poly-D-lysine (PDL) coated glass coverslips in 12-well plates at 24 hours prior to infection.
- Prepare aliquots of 150 μL of JUNV13 at a multiplication of infection (MOI) of 1.0 plaque forming unit per cell diluted in Dulbecco’s Modified Eagle’s Medium (DMEM) media supplemented with 2% fetal bovine serum (FBS) and 1% penicillin and streptomycin (P/S).
- Remove media from cell culture. Add virus inoculum on each well containing coverslip and incubate for 1.5 h at 37 °C. Shake the plates every 15 min.
- Remove the virus inoculum, add 1 mL of DMEM supplemented with 5% FBS and 1% P/S. Incubate the plate at 37 °C for desired time point.
2. Fixation and immunostaining
- Aspirate the media. Rinse the cells by adding 1 mL of phosphate buffered saline (PBS) supplemented with calcium and magnesium to each well.
- Remove PBS. Add 1 mL of methanol (MeOH) pre-chilled at -20 °C and incubate at -20 °C or on dry ice for 15 min.
- Remove MeOH.
- Add 1 mL of PBS to each well, and wash samples at 4 °C with gentle rocking for 5 min. Repeat the wash for total of 4 times.
- Wash the fixed cells on coverslips in 1 mL of 0.2% t-octylphenoxypolyethoxyethanol for 5 min at 4 °C, with gentle rocking.
- Add 1 mL of PBS to each well. Wash samples at 4 °C for 5 min with gentle rocking. Repeat the washing step for total of 4 times.
- To detect dsRNA and MDA5, incubate samples in 200 μL of primary antibodies diluted in 3% bovine serum albumin (BSA). Dilute the anti-dsRNA 9D5 antibody at a 1:2 dilution and dilute the anti-MDA-5 antibody at 1:250. Incubate with gentle rocking at 4 °C overnight.
- Remove primary antibodies and wash each well with 1 mL of PBS for 5 min with gentle rocking at RT. Repeat this wash four more times.
- Add 200 μL of secondary antibodies (1:2,000 dilution) diluted in 3% BSA and incubate at RT for 1 h.
- Remove secondary antibodies and wash each well with 1 mL of PBS for 5 min with gentle rocking at RT. Repeat this wash four more times.
- To detect JUNV nucleoprotein (NP) and RIG-I, add 200 μL of conjugated antibodies diluted in 3% BSA. Dilute the conjugated anti-JUNV NP (AG-12) at 1:1,000 and incubate samples for 2 h at RT with gentle rocking. Dilute the conjugated anti-RIG-I antibody at 1:500 and incubate at 4 °C overnight with gentle rocking.
- Remove conjugated antibodies and wash each well with 1 mL of PBS for 5 min with gentle rocking at RT. Repeat this wash four more times.
- Counterstain the coverslips with DAPI (1:1,000) for 3 min with gentle rocking at RT.
- Wash the coverslips 3 times, each time for 5 min in 1 mL of 0.5% t-octylphenoxypolyethoxyethanol with gentle rocking at RT.
- Wash twice in 1 mL of PBS for 5 min each time with gentle rocking at RT.
- Wash once in 1 mL of ddH2O for 1 min at RT, rocking gently.
- Mount coverslips onto glass slides using mounting media. Let cure overnight.
- Seal the slides with nail polish and air dry for 1 h.
- Image on confocal microscope with the 60x/1.42 numerical aperture oil immersion lens using the same laser emissions for each sample.
- When analyzing the data, if necessary, make adjustments for brightness and contrast using the same linear adjustment for all samples.
Representative Results
This protocol was applied to study the distribution and colocalization between the RLRs (RIG-I and MDA-5) and dsRNA in JUNV-infected cells. As shown in Figure 1 and Figure 2, the accumulation of dsRNA increases over time as viral infection progresses. Concentrated MDA-5 (Figure 1) and RIG-I (Figure 2) signals were found colocalized with the punctate structures of the NP and dsRNA.
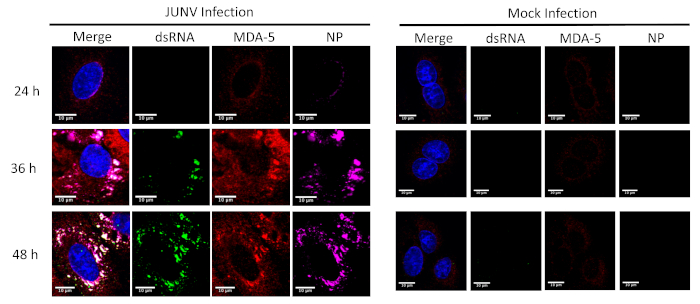
Figure 1: Time course of dsRNA and JUNV NP formation and the distribution of MDA-5. JUNV-infected and mock-infected A549 cells were fixed, stained and imaged according to the protocol at 24, 36, and 48 hours post infection (HPI). Please click here to view a larger version of this figure.
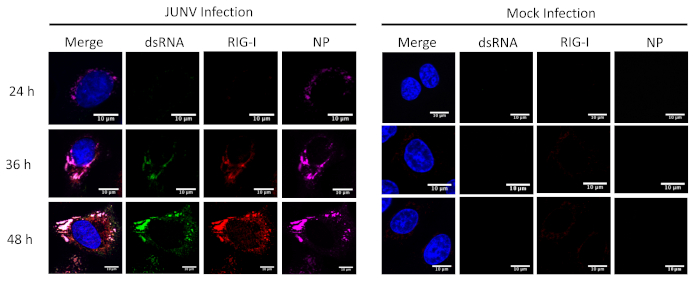
Figure 2: Time course of dsRNA and JUNV NP formation and the distribution of RIG-I. JUNV-infected and mock-infected A549 cells were fixed, stained and imaged according to the protocol at 24, 36, and 48 HPI. Please click here to view a larger version of this figure.
Discussion
For positive-sense RNA viruses and dsDNA viruses, dsRNA is readily detected with the widely-used J2 anti-dsRNA antibody. However, negative-sense RNA viruses are believed to produce dsRNA at a low or below the detection level using the same antibody2. Thus, many aspects of viral RNA and PRR interaction are largely unclear for negative-sense RNA viruses. We attempted to stain for dsRNA in arenavirus infection using the J2 antibody but the fluorescence signals were not differentiable compared to the mock infection. The Mab 9D5, originally used for pan-enterovirus detection, was found to be specific for dsRNA and more sensitive than the J2 antibody5. This antibody has been used successfully to detect viral dsRNA during negative-sense RNA virus infection, including the prototype arenavirus Lymphocytic choriomeningitis virus5,6,14. Accordingly, we used the Mab 9D5 to co-stain for the presence of dsRNA, the viral NP, and PRRs to further understand their distribution and interaction in individual cells during arenavirus infection.
There are multiple fixation methods that can be used to preserve cell structure. Fixation with MeOH acts by precipitating proteins, whereas paraformaldehyde and formalin crosslinks proteins. MeOH is more effective than aldehydes at conserving the nucleic acids in cells and provides low background immunostaining. Methanol also removes lipids from cells and thus permeabilizes cell membranes at the same time15. In this protocol, the cells are fixed using ice-cold MeOH. Other fixation methods were also attempted, including fixing samples with 4% paraformaldehyde, formalin, paraformaldehyde followed by methanol, and methanol followed by paraformaldehyde. However, high basal level non-specific staining was observed in mock-infected cells when paraformaldehyde or formalin was used. The optimal fixation method is fixation with MeOH at -20 °C based on the sensitivity and specificity of the results. Before primary antibody staining, sample blocking with 3% BSA, 5% BSA, and 10% or 5% goat serum for 30 min to 1 hour was tested, but all resulted in non-specific, background staining. The best results were achieved without the blocking step and directly using 3% BSA in the antibody dilution. The critical steps in minimizing the background signals are the PBS and t-Octylphenoxypolyethoxyethanol washes after fixation. While the commercially available 9D5 antibody is diluted by the vender and ready for direct use, a 1:2 dilution of the antibody with 3% BSA also worked well. In case that the dsRNA signal is weak, the antibody can be used without dilution.
This protocol can be utilized to study the interaction between dsRNA and PRRs in arenavirus infection. While this methodology was developed in a BSL-2 environment, the methanol fixation described herein allows complete inactivation of arenavirus. Therefore, the same protocol can be applied to imaging study for highly pathogenic arenaviruses (i.e., LASV, JUNV and MACV) in a BSL-4 facility. It is also possible to apply this protocol to studies on other RNA virus. To achieve optimal results, a modification of the protocol may be necessary.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We would like to thank the UTMB imaging core facilities and Maxim Ivannikov for microscope assistance.
This work was supported by Public Health Service grant RO1AI093445 and RO1AI129198 awarded to SP, UTMB Commitment Fund P84373 to CH, and T32 AI007526 to EM.
Materials
| APEX Alexa Fluor 647 antibody labeling kit | Invitrogen | A10475 | |
| BSA | Sigma Aldrich | A4503 | |
| DAPI | Cell Signaling | 4083 | 1:1,000 dilution |
| Donkey-anti rabbit Alexa Fluor 594 | Invitrogen | A-11058 | 1:2,000 dilution; Lot #: 1454437 |
| Dulbecco's modified Eagle's medium | Corning | 10-013-CV | |
| Fetal Bovine Serum | Atlanta Bio | S11150 | |
| Glass microscope slides | Fisher | 12-550-15 | |
| Goat-anti mouse Alexa Fluor 488 | Invitrogen | A-11029 | 1:2000 dilution; Lot #: 1874804 |
| Human lung epithelial A549 cells | ATCC | CCL-185 | |
| Methanol | Fisher | A412 | Stored at -20 °C |
| Mouse MAb anti-JUNV NP | BEI | NA05-AG12 | Conjugated to Alexa-647 at 1:1,000 dilution |
| Mouse MAb pan-Enterovirus 9D5 Reagent | Millipore Sigma | 3361 | ready for use; diluted to 1:2; Lot #: 3067445 |
| PBS supplemented with Ca and Mg | Corning | 21-030-CV | |
| PDL Coated coverslips | Neuvitro | H-12-1.5-pdl | |
| ProLong Gold antifade | Invitrogen | P10144 | |
| Rabbit MAb MDA-5 | Abcam | ab126630 | 1:250 dilution; Lot #: GR97758-7 |
| recombiant Candid#1 strain of JUNV | Lab generated | Lab generated | As previously described in reference 13. |
| RIG-I mouse MAb conjugated to Alexa-594 | Santa Cruz | sc-376845 | 1:1000 dilution; Lot #: AO218 |
| Triton X-100 | Sigma Aldrich | T8787 |
Referenzen
- Jensen, S., Thomsen, A. R. Sensing of RNA viruses: a review of innate immune receptors involved in recognizing RNA virus invasion. Journal of Virology. 86, 2900-2910 (2012).
- Weber, F., Wagner, V., Rasmussen, S. B., Hartmann, R., Paludan, S. R. Double-stranded RNA is produced by positive-strand RNA viruses and DNA viruses but not in detectable amounts by negative-strand RNA viruses. Journal of Virology. 80, 6 (2006).
- Triantafilou, K., Vakakis, E., Kar, S., Richer, E., Evans, G. L., Triantafilou, M. Visualisation of direct interaction of MDA5 and the dsRNA replicative intermediate form of positive strand RNA viruses. Journal of Cell Science. 125, 4761-4769 (2012).
- Onomoto, K., et al. Critical role of an antiviral stress granule containing RIG-I and PKR in viral detection and innate immunity. PLoS One. 7, e43031 (2012).
- Son, K. N., Liang, Z., Lipton, H. L. Double-stranded RNA is detected by immunofluorescence analysis in RNA and DNA virus infections, including those by negative-sense RNA viruses. Journal of Virology. 89, 9383-9392 (2015).
- Mateer, E. J., Paessler, S., Huang, C. Visualization of double-stranded RNA colocalizing with pattern recognition receptors in arenavirus infected cells. Frontiers Cellular and Infection Microbiology. 8, 251 (2018).
- Buchmeier, M. J., de la Torre, J. C., Peters, C. J., Knipe, D. M., Howley, P. M. Arenavirdae: The viruses and their replication. Fields Virology. 2, (2006).
- Levis, S. C., et al. Endogenous interferon in Argentine hemorrhagic fever. The Journal of Infectious Diseases. 149, 428-433 (1984).
- Levis, S. C., et al. Correlation between endogenous interferon and the clinical evolution of patients with Argentine hemorrhagic fever. Journal of Interferon Research. 5, 383-389 (1985).
- Huang, C., et al. Highly pathogenic new world and old world human arenaviruses induce distinct interferon response in human cells. Journal of Virology. 89, 7079-7088 (2015).
- Huang, C., et al. Junin virus infection activates the type I interferon pathway in a RIG-I-dependent manner. PLoS Neglected Tropical Diseases. 6, e1659 (2012).
- Huang, C., Kolokoltsova, O. A., Mateer, E. J., Koma, T., Paessler, S. Highly pathogenic new world arenavirus infection activates the pattern recognition receptor protein kinase R without attenuating virus replication in human cells. Journal of Virology. 91, 20 (2017).
- Emonet, S. F., et al. Rescue from cloned cDNAs and in vivo characterization of recombinant pathogenic Romero and live-attenuated Candid#1 strains of Junin virus, the causative agent of Argentine hemorrhagic fever disease. Journal of Virology. 85 (4), 1473-1483 (2011).
- Child, S. J., et al. Antagonism of the protein kinase R pathway in human cells by Rhesus Cytomegalovirus. Journal of Virology. 92, 6 (2018).
- Hobro, A. J., Smith, N. I. An evaluation of fixation methods: Spatial and compositional cellular changes observed by Raman imaging. Vibrational Spectroscopy. 91, 31-45 (2017).

