细菌中的RIBO-seq:NGS测序样本收集和库准备协议
Summary
在这里,我们描述了细菌中RIBO-seq样品收集和制备的阶段。根据这些准则对图书馆进行排序,可以获得足够的数据,以便进行全面的生物信息分析。我们目前的协议很简单,使用标准的实验室设备,从裂解到获得库需要七天时间。
Abstract
核糖体分析技术(RIBO-seq)是目前研究 体内蛋白质合成过程的最有效工具。与其他方法相比,这种方法的优点是它能够通过精确绘制 mRNA 成绩单上核糖体的位置和数量来监控翻译。
本文介绍了细菌中RIBO-seq方法的样品采集和制备的连续阶段,突出了与实验规划和执行相关的细节。
由于RIBO-seq依赖于完整的核糖体和相关的mRNA,关键步骤是快速抑制转化和细胞的充分解体。因此,我们建议在液氮中过滤和闪光冷冻,用于细胞采集,并可选使用氯霉素进行预处理,以阻止细菌的转化。对于解体,我们建议用砂浆研磨冷冻细胞,在氧化铝的存在下缠缠,以机械地破坏细胞壁。在此协议中,不需要蔗糖垫或蔗糖梯度超中心化用于单体净化。相反,使用聚丙烯酰胺凝胶电泳 (PAGE) 进行 mRNA 分离,然后应用核糖体足迹切除 (28-30 nt 波段),并提供令人满意的结果。这在很大程度上简化了方法,并减少了程序的时间和设备要求。为了准备图书馆,我们建议使用市售的小型RNA套件,用于新英格兰生物实验室的照明测序,遵循制造商的指南,并进行一定程度的优化。
由此产生的 cDNA 库为下一代测序 (NGS) 提出了所需的适当数量和质量。根据上述协议对准备的库进行排序,每个样本可绘制 2 到 10 毫升的独特映射读数,为全面的生物信息分析提供足够的数据。我们介绍的协议是快速和相对容易的,可以用标准的实验室设备执行。
Introduction
核糖体分析技术(RIBO-seq)是在加州大学旧金山分校乔纳森·魏斯曼的实验室里开发的。与用于研究转化水平基因表达的其他方法相比,RIBO-seq 专注于与 mRNA 结合的每个核糖体,并提供有关其位置和成绩单上核糖体的相对数量的信息。它能够监测体内蛋白质合成的过程,并可以提供单一的圆环分辨率和精度,从而能够测量细胞中单个 mRNA 和整个转录组的核糖体密度。RIBO-seq 技术的基础在于,在翻译过程中,核糖体将 mRNA 分子结合,从而保护脚本的埋藏片段免受核糖化的消化。加入核糖这些片段称为核糖体脚印(RF),然后可以分离、测序并映射到它们产生的脚本上,从而检测核糖体的确切位置。事实上,自20世纪60年代以来,保护mRNA片段的核糖体能力一直被用来研究核糖体结合和翻译启动站点(TIS)2,3,4。然而,随着深测序技术的进步,RIBO-seq已成为翻译监测5的黄金标准,通过核糖体的参与,可以提供蛋白质合成6的全基因组信息。核糖体分析填补了抄本组和原型6号之间存在的技术空白。
要进行核糖体分析,我们需要获得在调查条件下生长的生物体的细胞裂解。在细胞收集和裂解过程中破坏这些条件可能会提供不可靠的数据。为了防止这种情况,通常使用液氮中的转化抑制剂、快速收获和闪光冷冻。细胞可以通过机械同质化器(如搅拌机7、8或珠子搅拌机9)中的低温研磨来解冻,也可以通过移液器10或针11进行三次搅拌。裂解缓冲器可以在细胞粉碎之前或之后添加。在我们的协议中,我们使用液氮来预冷砂浆和虫害,以及氧化铝作为一种更温和的方法来破坏细菌细胞壁,防止RNA剪切经常遇到,当方法,如声化应用。粉碎后,我们将冰冷裂解缓冲器添加到砂浆的冷却内容中。选择适当的裂解缓冲区对于获得核糖体足迹的最佳分辨率非常重要。由于离子强度影响RF大小和读数帧精度,目前建议使用低离子强度和缓冲容量的裂解缓冲器,即使缓冲成分似乎不影响mRNA11,12的核糖体占用率。裂解缓冲的重要成分是镁离子,其存在可防止核糖核酸亚单位的分离,并抑制细菌核糖体11、13的自发构象变化。钙离子也起着重要作用,对于细菌核糖核酸(MNase)在细菌核糖体分析方法14中使用的活性至关重要。添加瓜诺辛5×[β,γ-米多]三磷酸盐(GMP-PNP),GTP的不可水解的模拟,以及氯霉素抑制在裂解15期间的翻译。
获得裂解剂时,通过离心机进行澄清,并分为两部分,每部分用于 RIBO-seq 和高通量总 mRNA 测序 (RNA-seq),因为它们同时执行 (图 1)。RNA-seq 提供了一个参考点,可在数据分析期间比较来自 RIBO-seq 和 RNA-seq 的数据。被调查的转子是由核糖体足迹正常化到mRNA丰度16来定义的。来自RNA-seq的数据也有助于识别克隆或测序文物17。
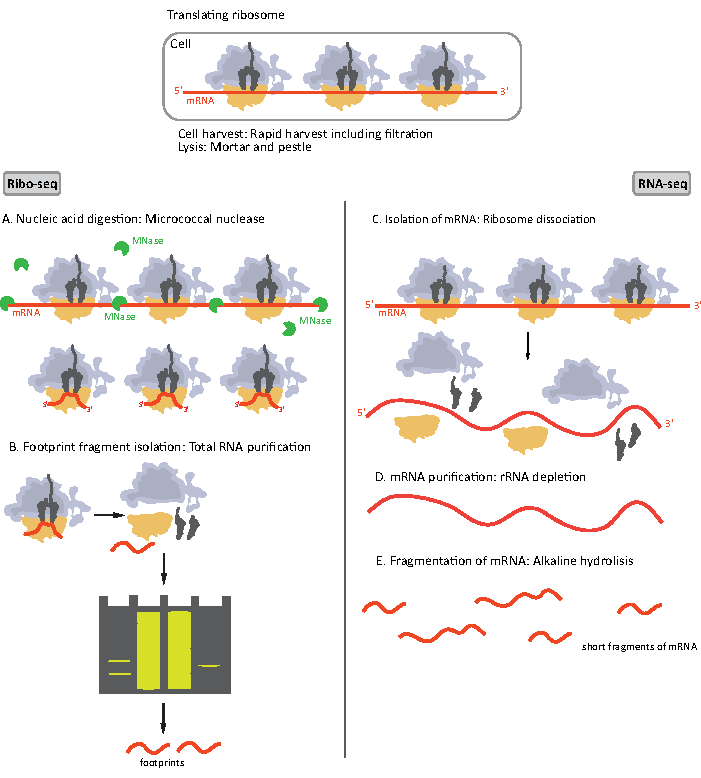
图1。为RIBO-seq和RNA-seq准备的mRNA样本的原理图。 对于 RIBO-seq 库的准备,RNA 会用 MNase (A) 进行消化,然后选择长度为 ~28-30 nt 长度的 RF 大小(B);RNA-seq RNA 是孤立的 (C),耗尽了 rRNA (D),由此产生的 mRNA 被随机分割成不同长度的片段 (E)。 请单击此处查看此图的较大版本。
RIBO-seq 和 RNA-seq 的样品制备程序的初始步骤略有不同(图 1)。对于核糖体分析,裂解酶需要由特定的内毒酶消化,以降解不受核糖体保护的mRNA分子。在标准协议中,获得的单体通过蔗糖垫超中心化或蔗糖梯度超中心化8,14回收。在本文中,我们表明,这一步骤是没有必要隔离的RF所需的RIBO-seq细菌,同样为真核细胞18,和大小选择适当的长度mRNA片段从多晶酰胺凝胶是足够的。
对于RNA-seq,mRNA是通过从总RNA-rRNA分子中耗尽到生物基化寡核苷酸探头获得的,后者与链球菌涂层的磁珠结合。然后用磁铁从样品中取出rRNA-寡核苷酸-珠复合物,导致rRNA耗尽样本19,20。纯化的mRNA分子然后随机碎片碱性水解。获得的 mRNA 片段以及核骨足迹被转换为 cDNA 库,并准备进行深测序(图 2)。这涉及碱性水解(用于 mRNA)和酶消化(用于 RF)后所需的末端修复:3′ 端的脱磷,然后是 5′ 端的磷酸化。接下来的步骤是适配器配合和反向转录,以创建 cDNA 插入,这些插入由使用 Illlumina 平台的下一代测序 (NGS) 所需的序列所示。库准备的最后阶段是 PCR 反应,其中结构被放大并贴上样品特定条形码的标签,以便在一个通道上对各种样品进行复用和排序。测序前,通过芯片电泳高灵敏度DNA评估库的质量和数量。然后,可以汇集和排序具有适当参数的 cDNA 库。可在不同的光照平台上进行排序,如 MiSeq、NextSeq 或 HighSeq,具体取决于库的数量、所需的读取长度和测序深度。测序后,进行生物信息分析。

图2。图书馆准备。 库准备包括末端修复、适配器结块、反向转录和用条形码放大。 请单击此处查看此图的较大版本。
核糖体剖析是一种通用的方法,可以根据科学问题进行易于修改和调整。最初它被用于酵母1,但不久后,它被应用于细菌细胞21以及真核模型生物,包括小鼠10,斑马鱼22,果蝇23和阿拉伯多普西塔利亚纳24。它也用于研究不同的核糖体类型:细胞质,线粒体25,26和叶绿体27,28。在欧盟核生物RIBO-seq是常见的适应和改进,以调查翻译的具体方面,包括启动10,11,29,30,31,32,拉长1,10,11,31,33,核糖体停滞33和构象变化33。大多数修改涉及使用不同的翻译抑制剂。然而,在细菌中,类似的研究一直难以进行,因为缺乏与行动34所需的机制的抑制剂。细菌中最常用的转化抑制剂是氯霉素 (CAM),它与肽转移酶中心 (PTC) 结合,防止氨基-tRNA在 A 站点的正确定位。因此,CAM阻止形成肽键,导致逮捕拉长核糖体35。细菌中翻译抑制剂的其他示例是四环素 (TET)36、雷塔帕穆林 (RET)34和 Onc11237,它们已用于调查翻译启动站点。TET,通过直接与A-site tRNA的抗子茎环重叠来防止tRNA输送到核糖体,最初用于验证从CAM治疗获得的结果,因为它们都是抑制翻译拉长38的抗生素。TET被发现检测原发性TIS,但无法显示内部TIS36。RET 结合细菌核糖体的 PTC,并通过干扰 A 站点中的拉长器氨基-tRNA 来防止形成第一个肽键。应用 RET 结果在小学和内部 TIS34中都会导致核糖体逮捕。Onc112,一种富含丙线的抗菌肽,在出口隧道中结合,并阻断核糖体A部位的氨基酸-tRNA结合。因此,Onc112 阻止启动复合物进入拉长阶段37。
核糖体分析提供的主要信息是核糖体密度及其在mRNA上的位置。它成功地用于研究不同生长条件下的翻译水平上的微分基因表达1、6、测量转化效率1、38、39,并检测利博索马尔暂停10等翻译调控事件。RIBO-seq还允许发现注释的ncRNA,伪基因和未注释的小开放读取帧(ORF)的翻译,导致识别新颖和/或非常短的蛋白质编码基因10,12,22,30,37。在这种情况下,RIBO-seq 可以微调和改进基因组注释。由于它对翻译的ORF的识别具有很高的敏感性和定量性质,核糖体分析也可以作为蛋白质组测定或辅助蛋白质组学研究31,34,39的代理。通过映射TIS,核糖体特征分析揭示了已知蛋白质10、32的N-末端延伸和截断等形。RIBO-seq也被改编为研究蛋白质14,21,24的共转化折叠。这种方法能够测量个体胶子6的拉长速率1、10、39或解码速度,并有助于开发翻译17的定量模型。核糖体分析方法还能够提供机械的见解,在细菌7,15,17,帧移位40,停止科顿读数21,终止/回收缺陷41,42和核糖体构象变化33在真核生物。RIBO-seq还被调整,以研究特定的转作用因子对翻译的影响,如miRNA6和RNA结合蛋白在真核生物16,43。然而,重要的是要承认,实验设计和获得的RIBO-seq分辨率决定了从由此产生的测序数据12中可以提取的信息量。
Protocol
Representative Results
Discussion
核糖体分析的关键技术挑战是需要快速抑制翻译,以便在特定的生理兴趣状态下捕获 mRNA 上的核糖体快照。为此,通常使用液氮中的转化抑制剂、快速收获和闪光冷冻。使用抗生素是可选的,因为它们会导致人工制品。氯霉素是一种常用的药物,用于抑制细菌RIBO-seq中拉长的核糖体。然而,它并不阻止启动,导致核糖体的积累约6个科顿下游的翻译启动网站14。此外,其抑制肽转?…
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
ALS 感谢 EMBO 安装赠款 IG 3914 和 POIR 的财政支持。04.04.00-00-3E9C/17-00 在欧洲联盟根据欧洲区域发展基金共同资助的波兰科学基金会第一小组方案内进行。
Materials
| 10X TBE (powder) | Invitrogen | AM9864 | |
| 2-Mercaptoethanol, 99%, pure | Acros Organics | 125472500 | |
| Adenosine 5'-Triphosphate (ATP) | New England Biolabs | P0756S | |
| Aluminium oxide calcinated pure p.a. | Chempur | 114560600 | |
| Calcium chloride dihydrate | Sigma-Aldrich | C3881-500G | |
| Chloramphenicol | MP Biomedicals | 190321 | |
| DNA Clean & Concentrator -5 | Zymo Research | D4004 | |
| Dnase I recombinant, Rnase-free | Roche | 4716728001 | |
| EDTA disodium salt | Fisher Scientific | E/P140/48 | |
| Ethyl Alcohol Absolut 99,8% Pure-P.A.-Basic | POCH Avantor Performance Materials Poland S.A | BA6480111 | |
| Filtration apparatus | VWR Collection | 511-0265 | all-glass filtration apparatus, with funnel, fritted base, cap, 47 mm Ø spring clamp and ground joint flask |
| Gel 40 (19:1) | Rotiphorese | 3030.1 | |
| Gel Loading Dye, Blue, 6X | New England Biolabs | E6138G | |
| Guanosine 5′-[β,γ-imido]triphosphate trisodium salt hydrate | Sigma-Aldrich | G0635-25MG | |
| labZAP | A&A Biotechnology | 040-500 | |
| Magnesium acetate tetrahydrate | Sigma-Aldrich | M5661-250G | |
| MCE membrane fiter | Alfatec Technology | M47MCE45GWS | pore size: 0.45um |
| MICROBExpress Bacterial mRNA Purification | Invitrogen | AM1905 | |
| Multiplex Small RNA Library Prep Set for Illumina | New England Biolabs | E7300S | |
| Nuclease-Free Water | Ambion | AM9937 | |
| Potassium Acetate Anhydrous Pure P.A. | POCH Avantor Performance Materials Poland S.A | 744330113 | |
| Quick-Load pBR322 DNA-MspI Digest | New England Biolabs | E7323A | |
| RNA Clean & Concentrator -25 | Zymo Research | R1018 | |
| Sodium acetate | Sigma-Aldrich | S2889-250G | |
| Sodium carbonate | Sigma-Aldrich | 223530-500G | |
| Sodium hydrogen carbonate pure p.a. | POCH Avantor Performance Materials Poland S.A | 810530115 | |
| SYBR Gold nucleic acid gel stain | Life Technologies | S11494 | |
| T4 Polynucleotide Kinase | New England Biolabs | M0201L | |
| T4 Polynucleotide Kinase Reaction Buffer | New England Biolabs | B0201S | |
| TBE-Urea Sample Buffer (2x) | Invitrogen | LC6876 | |
| Tris(hydroxymethyl)amino-methane, ultrapure, 99,9% | AlfaAesar | J65594 | |
| Triton X-100, 98% | Acros Organics | 327371000 | |
| Urea G.R. | lach:ner | 40096-AP0 |
Referenzen
- Ingolia, N. T., Ghaemmaghami, S., Newman, J. R., Weissman, J. S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 324 (5924), 218-223 (2009).
- Takanami, M., Yan, Y., Jukes, T. H. Studies on the site of ribosomal binding of f2 bacteriophage RNA. Journal of Molecular Biology. 12 (3), 761-773 (1965).
- Steitz, J. A. Polypeptide Chain Initiation: Nucleotide Sequences of the Three Ribosomal Binding Sites in Bacteriophage R17 RNA. Nature. 224 (5223), 957-964 (1969).
- Wolin, S. L., Walter, P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. The EMBO journal. 7 (11), 3559-3569 (1988).
- Argüello, R. J., et al. SunRiSE – measuring translation elongation at single-cell resolution by means of flow cytometry. Journal of Cell Science. 131 (10), 214346 (2018).
- Michel, A. M., Baranov, P. V. Ribosome profiling: a Hi-Def monitor for protein synthesis at the genome-wide scale. Wiley Interdisciplinary Reviews: RNA. 4 (5), 473-490 (2013).
- Woolstenhulme, C. J., Guydosh, N. R., Green, R., Buskirk, A. R. High-precision analysis of translational pausing by ribosome profiling in bacteria lacking EFP. Cell reports. 11 (1), 13-21 (2015).
- McGlincy, N. J., Ingolia, N. T. Transcriptome-wide measurement of translation by ribosome profiling. Methods (San Diego, Calif). 126, 112-129 (2017).
- Gerashchenko, M. V., Gladyshev, V. N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic acids research. 42 (17), 134 (2014).
- Ingolia, N. T., Lareau, L. F., Weissman, J. S. Ribosome Profiling of Mouse Embryonic Stem Cells Reveals the Complexity of Mammalian Proteomes. Cell. 147 (4), 789-802 (2011).
- Ingolia, N. T., Brar, G. A., Rouskin, S., McGeachy, A. M., Weissman, J. S. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nature protocols. 7 (8), 1534-1550 (2012).
- Hsu, P. Y., et al. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America. 113 (45), 7126-7135 (2016).
- Blanchard, S. C., Kim, H. D., Gonzalez, R. L., Puglisi, J. D., Chu, S. tRNA dynamics on the ribosome during translation. Proceedings of the National Academy of Sciences of the United States of America. 101 (35), 12893-12898 (2004).
- Becker, A. H., Oh, E., Weissman, J. S., Kramer, G., Bukau, B. Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nature protocols. 8 (11), 2212-2239 (2013).
- Li, G. W., Oh, E., Weissman, J. S. The anti-Shine-Dalgarno sequence drives translational pausing and codon choice in bacteria. Nature. 484 (7395), 538-541 (2012).
- King, H. A., Gerber, A. P. Translatome profiling: methods for genome-scale analysis of mRNA translation. Briefings in functional genomics. 15 (1), 22-31 (2014).
- Mohammad, F., Woolstenhulme, C. J., Green, R., Buskirk, A. R. Clarifying the Translational Pausing Landscape in Bacteria by Ribosome Profiling. Cell reports. 14 (4), 686-694 (2016).
- Reid, D. W., Shenolikar, S., Nicchitta, C. V. Simple and inexpensive ribosome profiling analysis of mRNA translation. Methods (San Diego, Calif). 91, 69-74 (2015).
- Sanz, E., Yang, L., Su, T., Morris, D. R., McKnight, G. S., Amieux, P. S. Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proceedings of the National Academy of Sciences of the United States of America. 106 (33), 13939-13944 (2009).
- Petrova, O. E., Garcia-Alcalde, F., Zampaloni, C., Sauer, K. Comparative evaluation of rRNA depletion procedures for the improved analysis of bacterialbiofilm and mixed pathogen culture transcriptomes. Scientific Reports. 7, 41114 (2017).
- Oh, E. Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell. 147 (6), 1295-1308 (2011).
- Chew, G. L. Ribosome profiling reveals resemblance between long non-coding RNAs and 5′ leaders of coding RNAs. Development. 140 (13), 2828-2834 (2013).
- Dunn, J. G., Foo, C. K., Belletier, N. G., Gavis, E. R., Weissman, J. S. Ribosome profiling reveals pervasive and regulated stop codon readthrough in Drosophila melanogaster. eLife. 2, 01179 (2013).
- Liu, M. J., et al. Translational landscape of photomorphogenic Arabidopsis. The Plant cell. 25 (10), 3699-3710 (2013).
- Rooijers, K., Loayza-Puch, F., Nijtmans, L. G., Agami, R. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nature Communications. 4, 2886 (2013).
- Danielle, L., et al. Fidelity of translation initiation is required for coordinated respiratory complex assembly. Science Advances. 5 (12), 2118 (2019).
- Zoschke, R., Watkins, K. P., Barkan, A. A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. The Plant cell. 25 (6), 2265-2275 (2013).
- Chotewutmontri, P., Barkan, A. Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS genetics. 12 (7), 1006106 (2016).
- Lee, S., Liu, B., Huang, S. X., Shen, B., Qian, S. B. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proceedings of the National Academy of Sciences of the United States of America. 109 (37), 2424-2432 (2012).
- Martinez, T. F., Chu, Q., Donaldson, C., Tan, D., Shokhirev, M. N., Saghatelian, A. Accurate annotation of human protein-coding small open reading frames. Nature chemical biology. 16 (4), 458-468 (2020).
- Simsek, D., et al. The Mammalian Ribo-interactome Reveals Ribosome Functional Diversity and Heterogeneity. Cell. 169 (6), 1051-1065 (2017).
- Fritsch, C., et al. Genome-wide search for novel human uORFs and N-terminal protein extensions using ribosomal footprinting. Genome research. 22 (11), 2208-2218 (2012).
- Lareau, L. F., Hite, D. H., Hogan, G. J., Brown, P. O. Distinct stages of the translation elongation cycle revealed by sequencing ribosome-protected mRNA fragments. eLife. 3, 01257 (2014).
- Meydan, S., et al. Retapamulin-Assisted Ribosome Profiling Reveals the Alternative Bacterial Proteome. Molecular Cell. 74 (3), 481-493 (2019).
- Wilson, D. N. The A-Z of bacterial translation inhibitors. Critical Reviews in Biochemistry and Molecular Biology. 44, 393-433 (2009).
- Nakahigashi, K. Comprehensive identification of translation start sites by tetracycline-inhibited ribosome profiling. DNA research : an international journal for rapid publication of reports on genes and genomes. 23 (3), 193-201 (2016).
- Weaver, J., Mohammad, F., Buskirk, A. R., Storz, G. Identifying Small Proteins by Ribosome Profiling with Stalled Initiation Complexes. mBio. 10 (2), 02819 (2019).
- Nakahigashi, K. Effect of codon adaptation on codon-level and gene-level translation efficiency in vivo. BMC genomics. 15 (1), 1115 (2014).
- Li, G. W., Burkhardt, D., Gross, C., Weissman, J. S. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell. 157 (3), 624-635 (2014).
- Michel, A. M., et al. Observation of dually decoded regions of the human genome using ribosome profiling data. Genome research. 22 (11), 2219-2229 (2012).
- Guydosh, N. R., Green, R. Dom34 rescues ribosomes in 3′ untranslated regions. Cell. 156 (5), 950-962 (2014).
- Young, D. J., Guydosh, N. R., Zhang, F., Hinnebusch, A. G., Green, R. Rli1/ABCE1 Recycles Terminating Ribosomes and Controls Translation Reinitiation in 3’UTRs In Vivo. Cell. 162 (4), 872-884 (2015).
- Ingolia, N. T. Ribosome profiling: new views of translation, from single codons to genome scale. Nature Reviews Genetics. 15 (3), 205-213 (2014).
- Dobin, A., et al. ultrafast universal RNA seq aligner. Bioinformatics. 29 (1), 15-21 (2013).
- . . Babraham Bioinformatics. Fast QC. , (2021).
- Gerashchenko, M. V., Gladyshev, V. N. Ribonuclease selection for ribosome profiling. Nucleic acids research. 45 (2), 6 (2017).
- Verbruggen, S., Menschaert, G. mQC: A post-mapping data exploration tool for ribosome profiling. Computer Methods and Programs in Biomedicine. 181, 104806 (2019).
- Cui, H., Hu, H., Zeng, J., Chen, T. DeepShape: estimating isoform-level ribosome abundance and distribution with Ribo-seq data. BMC bioinformatics. 20, 678 (2019).
- Choi, J. . RiboToolkit: an integrated platform for analysis and annotation of ribosome profiling data to decode RNA translation at codon resolution. , (2021).
- Choi, J. Dynamics of the context-specific translation arrest by chloramphenicol and linezolid. Nature chemical biology. 16 (3), 310-317 (2020).
- Tompson, J., O’Connor, M., Mills, J. A., Dahlberg, A. E. The protein synthesis inhibitors, oxazolidinones and chloramphenicol, cause extensive translational inaccuracy in vivo. Journal of Molecular Biology. 322 (2), 273-279 (2002).
- Brar, G. A., Weissman, J. S. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nature reviews. Molecular cell biology. 16 (11), 651-664 (2015).
- Glaub, A., Huptas, C., Neuhaus, K., Ardern, Z. Recommendations for bacterial ribosome profiling experiments based on bioinformatic evaluation of published data. Journal of Biological Chemistry. 295, 8999-9011 (2020).
- Sorour, M. H., Hani, H. A., Shaalan, H. F., El-Sayed, M. M. H. Experimental screening of some chelating agents for calcium and magnesium removal from saline solutions. Desalination and Water Treatment. 57 (48-49), 22799-22808 (2015).
- Mohammad, F., Green, R., Buskirk, A. R. A systematically-revised ribosome profiling method for bacteria reveals pauses at single-codon resolution. Elife. 8, 42591 (2019).
- O’Connor, P. B., Li, G. W., Weissman, J. S., Atkins, J. F., Baranov, P. V. rRNA:mRNA pairing alters the length and the symmetry of mRNA-protected fragments in ribosome profiling experiments. Bioinformatics. 29 (12), 1488-1491 (2013).
- Protocol for RNA Clean & Concentrator -25. Zymo Research Available from: https://files.zymoresearch.com/protocols/_r1017_r1018_rna_clean_concentrator-25.pdf (2021)
- Protocol for use with NEBNext Small RNA Library Prep Set for Illumina (E7300, E7580, E7560, E7330). New England Biolabs Available from: https://www.international.neb.com/protocols/2018/03/27/protocol-for-use-with-nebnext-small-rna-library-prep-set-for-illumina-e7300-e7580-e7560-e7330 (2021)

