Generation and Downstream Analysis of Single-Cell and Single-Nuclei Transcriptomes in Brain Organoids
Summary
Here, we introduce a comprehensive protocol for the generation and downstream analysis of human brain organoids using single-cell and single-nucleus RNA sequencing.
Abstract
Over the past decade, single-cell transcriptomics has significantly evolved and become a standard laboratory method for simultaneous analysis of gene expression profiles of individual cells, allowing the capture of cellular diversity. In order to overcome limitations posed by difficult-to-isolate cell types, an alternative approach aiming at recovering single nuclei instead of intact cells can be utilized for sequencing, making transcriptome profiling of individual cells universally applicable. These techniques have become a cornerstone in the study of brain organoids, establishing them as models of the developing human brain. Leveraging the potential of single-cell and single-nucleus transcriptomics in brain organoid research, this protocol presents a step-by-step guide encompassing key procedures such as organoid dissociation, single-cell or nuclei isolation, library preparation and sequencing. By implementing these alternative approaches, researchers can obtain high-quality datasets, enabling the identification of neuronal and non-neuronal cell types, gene expression profiles, and cell lineage trajectories. This facilitates comprehensive investigations into cellular processes and molecular mechanisms shaping brain development.
Introduction
Over the last years, organoid technologies have emerged as a promising tool to culture organ-like tissues1,2,3. Especially for organs that cannot be easily accessed, such as the human brain, organoids offer the opportunity to gain insights into development and disease manifestation4. As such, brain organoids have been widely used as an experimental model to investigate various human brain disorders, including developmental, psychiatric, or even neurodegenerative diseases4,5,6.
With the advent of single-cell transcriptome profiling technologies, primary human tissues and complex in vitro models could be studied with an unprecedented level of granularity, providing mechanistic insights into gene expression changes on the level of cell subpopulations in health and disease and informing about new putative therapeutic targets7,8,9. The organoid field has progressed by utilizing single-cell transcriptome profiling to assess cellular composition, reproducibility and the fidelity of brain organoid technologies10,11,12. Single-cell RNA-sequencing (scRNA-seq) enabled cell classification and the identification of genetic dysregulation in diseased organoids13,14. Importantly, it is the complexity of organoid tissues that necessitates the implementation of techniques that enable the profiling of individual cells. Characterization of organoids using methods such as bulk transcriptome profiling (bulk RNA sequencing) leads to masked cellular heterogeneity and gene expression profiles which are averaged across all types of cells within the complex tissue, ultimately limiting our understanding of ongoing processes during organoid development in health and disease15,16,17. As scRNA-seq methods continue to advance, an increasing number of atlases are being created, exemplified by resources like the Allen Brain Atlas or the Single cell atlas of human brain organoids by Uzquiano et al.18.
Accomplishing successful scRNA-seq from brain organoids relies on effective isolation and capture of intact cells. As the dissociation of brain organoids to obtain individual cells is based on enzymatic digestion, it can influence gene expression patterns by inducing stress and cell damage19,20. Hence, the dissociation of the tissue into individual cells is the most crucial step. An alternative approach is single-nucleus RNA sequencing (snRNA-seq), which facilitates the enzyme-free extraction of nuclei from both, fresh and frozen, tissue21,22. However, the isolation of nuclei from a tissue poses other challenges such as the enrichment of cell types of interest and the low RNA content of nuclei in comparison to cells.
Transcriptome studies of brain organoids are commonly conducted using scRNA-seq10,18,23. However, the isolation of single nuclei might provide an orthogonal and supplemental method to investigate the transcriptomic profile of organoids. Here, we introduce a toolbox for scRNA- and snRNA-seq for brain organoids and discuss the critical points for obtaining the best quality sequencing data.
Protocol
The described protocol is performed in a biosafety level 1 laboratory of the Max Delbrück Center for Molecular Medicine (approval number: 138/08), in accordance with the requirements and in compliance with EU and national rules on ethics in research.
1. Derivation of forebrain organoids from induced pluripotent stem cells (iPSCs)
NOTE: This protocol was tested for several different iPSC lines cultured in a variety of stem cell media from different companies (Table 1). The generation of forebrain organoids is highly dependent on high quality iPSCs and a confluence of 60%-70% prior to starting the protocol. Here we used a commercially available cell line (see Table of Materials).
- Day 0: Embryoid body formation
- For treatment of 96-well plate with pluronic solution, add 80 µL of sterile filtered 2% pluronic solution per well of a 96-well U-bottom plate. Incubate for at least 15 min at room temperature (RT) or 37 °C.
NOTE: Pluronic treated plates can be stored at 4 °C for up to 2 weeks and can be used directly without any washing steps for embryoid body formation. - Remove pluronic solution from each well using an aspirator or a multichannel pipette before seeding cells.
- Before starting, prepare the following reagents for one 96-well plate: 15 mL centrifuge tube with 5 mL of pre-warmed DMEM: F12 medium; 11 mL of stem cell medium supplemented with 50 µM Y-27632, pluronic-treated plate (as described above).
- Aspirate media from one well of a 6-well plate of 60-70% confluent iPSCs. Wash cells once with PBS. Add 500 µL of trypsin-based reagent and incubate for 2-6 min at 37 °C.
NOTE: Incubation time for proper detachment of cells with trypsin-based reagent depends on cell density and the cell matrix-adhesive properties of each iPSC line. To avoid over digestion, it is advised to examine cell morphology after 2 min of incubation with trypsin-based reagent using a bright field microscope. - Detach cells using a 1000 µL pipette and transfer into the previously prepared 15 mL centrifuge tube with DMEM:F12 medium to stop dissociation.
- Centrifuge cell suspension for 3 min at 300 x g. Aspirate supernatant and keep the cell pellet. Resuspend the cells in 1 mL of stem cell medium supplemented with 50 µM Y-27632.
- Count cells using a cell counter and add desired number of cells to the stem cell media supplemented with 50 µM Y-27632. The generation of a single embryoid body requires 3,000-12,000 cells depending on the cell line.
- Transfer the cell suspension to a multichannel reagent reservoir using a 10 mL pipette. Using a multichannel pipette, plate 100 µL/well of the cell suspension into the previously pluronic-treated 96-well plate.
- Place 96-well plate in an incubator at 37 °C, 5% O2 and 5% CO2. To avoid any disturbance during embryoid body formation, refrain from moving the plate.
- For treatment of 96-well plate with pluronic solution, add 80 µL of sterile filtered 2% pluronic solution per well of a 96-well U-bottom plate. Incubate for at least 15 min at room temperature (RT) or 37 °C.
- Day 2: Embryoid body formation examination
- Examine embryoid body formation using a 10x objective of a bright field microscope. Embryoid bodies should display clear edges and minimal cell death.
- Aspirate as much medium as possible and replace with 100 µL/well of stem cell medium. (Critical) Embryoid bodies are easily removed from the well during medium exchange. Pay attention to not remove them, therefore monitor medium after aspiration.
- Day 3: Neuroectoderm induction
- Aspirate as much medium as possible and replace with 120 µL/well of induction medium and place plate at 37 °C, 20% O2 and 5% CO2.
- Day 6: Embedding
- Depending on the iPSC line, 3 or 4 days are required to induce neuroectoderm emergence. Monitor embryoid bodies regularly. Once the edges turn bright, embryoid bodies are ready for embedding. Use one of the two different methods that can be used for embryoid body embedding as described below24.
- Embedding method 1: Cookie embedding
- Thaw an aliquot of undiluted extracellular matrix gel on ice for 1-2 h. (Critical) Always keep extracellular matrix gel on ice to prevent it from solidifying. Prepare aliquots in advance to minimize thawing time and repeated thawing cycles.
- Cut the tip of a 200 µL pipette using claw nippers and collect 16-32 embryoid bodies in one 1.5 mL microcentrifuge tube.
- Transfer the embryoid bodies in 67 µL of the media into a new 1.5 mL microcentrifuge tube. Add 100 µL of extracellular matrix gel using a cut 200 µL pipette tip and mix gently.
- Evenly distribute the embryoid bodies in a 6 cm dish and place the plate for 5-10 min to allow the extracellular matrix gel to solidify.
- Add about 4 mL of differentiation media and incubate at 37 °C, 20% O2 and 5% CO2. On the next day, place the dish on an orbital shaker (84 rpm). Ensure that all embryoid bodies detach from the bottom. If not, use a cut pipette tip and media to gently detach them.
- Embedding method 2: Liquid embedding
- Thaw an aliquot of undiluted extracellular matrix gel on ice for 1-2 h and place differentiation media on ice. (Critical) Media must be ice-cold when adding extracellular matrix gel.
- Once the medium is cold, add extracellular matrix gel to the final concentration of 2%. Cut the tip of a 200 µL pipette using claw nippers and transfer up to 24 embryoid bodies to one well of a 6-well plate.
- Remove all excess media and add about 4 mL of 2% extracellular matrix gel/differentiation media into one well of a 6-well plate. Immediately place the plate on the orbital shaker (84 rpm).
- Day 9-20: Perform a full media exchange using differentiation media every 3-4 days.
- Day 21-30: Transfer organoids into maturation media and perform full media exchanges every 3-4 days.
- Dissociation of organoids: For single nuclei and cell dissociation, use organoids of high quality. To avoid excess amounts of cell death, regularly cut the organoids to prevent the build-up of a necrotic core and allow them to recover for 2 weeks before dissociating them for further analysis.
2. Derivation of single cell from organoids
NOTE: Single cell dissociation is performed using the Neural Tissue Dissociation Kit (Table 2), which uses mechanical and enzymatic dissociation. Here we describe a manual mechanical dissociation. As an alternative, a dissociation machine can be used.
- Prepare STOP solution and 0.04% BSA on ice. Prepare enzyme mix 1 and 2 and 0.04% BSA in PBS and place on ice.
- Collect up to 5 organoids in one well of a 6-well plate and aspirate excess media and wash once with PBS to remove culture media. Place organoids in the middle of the well and using a scalpel, mince organoids thoroughly.
- Add enzyme mix 1 and place at 37 °C, 20% O2 and 5% CO2, shaking at 90 rpm for 10-15 min.
- Using a 1000 µL pipette tip, resuspend organoids by pipetting up to 10x. Add enzyme mix 2 and mix it well by pipetting using a 1000 µL pipette tip. Place at 37 °C, 20% O2 and 5% CO2, shaking at 90 rpm for 10-15 min.
- Stop the dissociation process by resuspending the cell solution in 10 mL of cold STOP solution. Filter cell solution using a 70 µM cell strainer. Centrifuge the filtered cell solution for 10 min at 300 x g at 4 °C to form a pellet.
- Aspirate media, leaving a cell pellet and resuspend cells in 4 mL of cold 0.04% BSA. Filter cell solution using a 40 µM cell strainer and count cells using a cell counter.
- Centrifuge the cell solution for 10 min at 300 x g at 4 °C. Aspirate BSA solution, leaving a cell pellet. Resuspend cells according to a microfluidics-based-snRNA-seq kit manual in NSB+ medium.
3. Isolation of single nuclei from organoids
- Transfer organoids into chilled PBS and cut each organoid into 4 pieces. Wash organoids 2x with chilled PBS.
- Cut with scissors the pipette tip of a 1000 µL pipette and transfer organoid pieces into a 2 mL douncer. Aspirate PBS completely and add 1 mL of NP40 lysis buffer.
- Dounce 3x with dounce pestle A and 3x with dounce pestle B. Transfer suspension into a 15 mL centrifuge tube. Wash douncer with an additional 1 mL of NP40 lysis buffer and transfer into a 15 mL centrifuge tube. Incubate for 5 min at RT and subsequently centrifuge suspension for 5 min at 500 x g.
- Aspirate supernatant leaving 50 µL of supernatant behind. Add 1 mL of NSB+ without disturbing the pellet. Resuspend after 5 min of incubation.
- Layer suspension on top of a Percoll gradient (bottom layer: 1 mL of NSB + and 250 µL of Percoll, middle layer: 1 mL of NSB+ and 188 µL of Percoll, top layer: 1 mL of NSB+ and 125 µL of Percoll). (Critical) Perform layering very carefully to avoid layer mixing.
- Centrifuge for 5 min at 500 x g at 4 °C, aspirate supernatant and resuspend in NSB+. Filter nuclei solution using a 40 µM cell strainer.
- Stain and count nuclei using DAPI and a Neubauer Chamber. Resuspend nuclei according to a microfluidics-based scRNA-seq kit manual.
4. Library preparation and sequencing
- Load 10,000 cells and nuclei into a microfluidics system and generate the library according to the manual of microfluidics based scRNA-seq kit (Table of Materials).
- Sequence the libraries with an estimated 6 x 108 reads per snRNA-seq library and 1.6 x 108 reads per scRNA-seq library, resulting in a sequencing saturation of 87% for the snRNA-seq dataset and 36% sequencing saturation for the scRNA-seq dataset.
5. Analysis
- Carry out analysis of sequencing using a data analysis pipeline for scRNA-seq and snRNA-seq and align against the human GRCh38 genome. Subject the count matrix generated by this pipeline to further analysis using the R-package Seurat (Version 4.1.1)25.
- Create the Seurat object by considering genes that were represented in at least 5 cells and incorporating cells that contained at least 300 genes. Perform additional quality control filtering by retaining cells that contained more than 1000 genes but less than 4000 genes for the scRNA-seq dataset and more than 800, but less than 4000 genes per nuclei for the snRNA-seq dataset. Exclude datasets cells and nuclei exceeding 5% mitochondrial reads.
- To mitigate batch effects between both methods, integrate both filtered datasets, normalize and visualize via Uniform Manifold Approximation and Projection (UMAP). Omit cluster 1 which displays low unique molecular identifier (UMI) and gene counts. Afterwards, for the clusters assign cell identities based on known marker genes and the 30-day-old organoid dataset from Ana Uzquiano et. al. (Supplementary Coding File 1)18.
Representative Results
To investigate cell type composition of brain organoids using scRNA-seq and snRNA-seq, brain organoids were harvested after 30 days of culture as organoids at this stage already exhibit neuroepithelial loops consisting of progenitors surrounded by intermediate progenitors and early stage neurons4,18. Monitoring the quality of the organoids throughout growth and culturing is essential for obtaining reliable single-cell and single-nucleus data.
Organoids are formed through the aggregation of iPSCs into embryoid bodies (Figure 1B,C). Upon assembly, these embryoid bodies are expected to exhibit clear edges with minimal cellular debris (Figure 1C). Following the induction of the neuroectoderm, the embryoid bodies manifest an observable brightening around the surface with a comparatively darker inner region, indicating successful neural induction (Figure 1D). In the following weeks, the organoids develop so-called loops, neuronal rosette-like structures, primarily composed of neuroepithelial cells (Figure 1E,F). Although these organoids can be maintained in culture over several months, it should be noted that with their increasing size, there is a corresponding increase of cell death in the inner part of the organoids due to the lack of nutrients and oxygen availability, eventually resulting in the development of a necrotic core.
In order to explore the cellular diversity within cortical organoids through sequencing analysis, we isolated both single cells and nuclei from organoids. To balance out the inherent heterogeneity within a single batch of brain organoids, we conducted sequencing of four pooled organoids from one batch. Additionally, for comparative purposes between the isolation processes and to remove batch effects between the snRNA-seq and the scRNA-seq library, these organoids were divided into halves (eight halves in total; Figure 2). Following the enzymatic isolation of individual cells, it is crucial to ensure that cell viability remains above 80%, while observing that the cells maintain a round morphology (Figure 3A,B). Similarly, the mechanical isolation of individual nuclei should yield intact nuclei of varying sizes with minimal to no detectable debris (Figure 3C,D).
Sequencing analysis revealed that more cells than nuclei were captured and sequenced. Both datasets showed a good quality assessed by the high gene and UMI count per cell and nucleus, and the low percentage of mitochondrial reads (Figure 4A-C). As expected, mitochondrial as well as ribosomal reads comprise a higher fraction of total reads recovered in the scRNA-seq dataset as compared to the snRNA-seq dataset. In the scRNA-seq dataset, most cells contained less than 30% ribosomal reads, whereas in the snRNA-seq dataset, the majority of nuclei contained less than 5% ribosomal reads. Additionally, the majority of cells captured within the scRNA-seq dataset contain less than 5% mitochondrial reads. After filtering out mitochondrial reads, around 10,000 cells and 3,000 nuclei passed the quality threshold.
Integrative analysis of both datasets revealed that all clusters are represented in both datasets, indicating both methods recover the same cell types (Figure 5A,C). The annotation of the dataset shows that the major cell populations comprise of radial glia, intermediate progenitors, subcortical progenitors and neurons, as well as newborn deep layer projection neurons and less represented cell types such as Cajal-Retzius, the cortical hem and choroid plexus cells (Figure 5B). Overall, scRNA-seq and snRNA-seq could recover all cell types expected to be present in a 30-day-old brain organoid20.
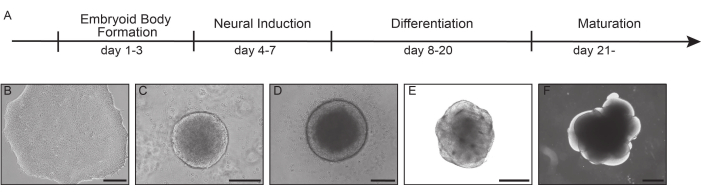
Figure 1: Generation of brain organoids from iPSCs. Time course of brain organoid generation (A). Exemplary images of a successful cortical differentiation from iPSCs (B), which form embryoid bodies (72 h after seeding) (C) and show a successful neural induction after 7 days of culture (D). Organoid depicts distinct loops at day 15 (E) and day 30 (F). Scale bar: B-D 200 µm, E 400 µm, F 800 µm. Please click here to view a larger version of this figure.
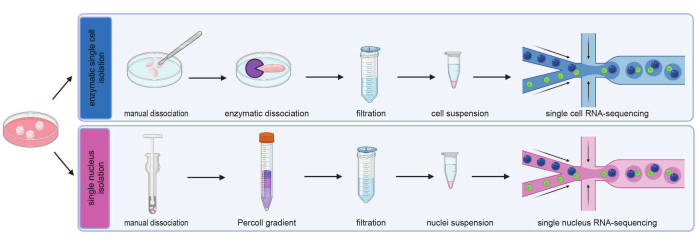
Figure 2: Workflow to obtain single cells and single nuclei from brain organoids. Dissociation of organoids into single cells is conducted by mincing organoids with a scalpel, followed by an enzymatic digestion (top). Isolation of nuclei is conducted by mechanical dissociation, followed by purification via a Percoll gradient (bottom). The single cell and nuclei suspension are filtered and loaded into a microfluidics system for library generation and sequencing. Please click here to view a larger version of this figure.
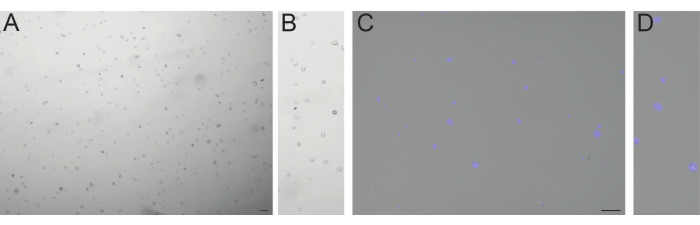
Figure 3: Representative results of isolated cells and nuclei from 30-day-old organoids. (A) Exemplary picture of trypan blue-stained cells taken using a cell counter and (B) corresponding close-up. (C) Overlay of brightfield and fluorescent picture of DAPI stained nuclei and (D) corresponding close-up. Scale bar: 100 µM Please click here to view a larger version of this figure.
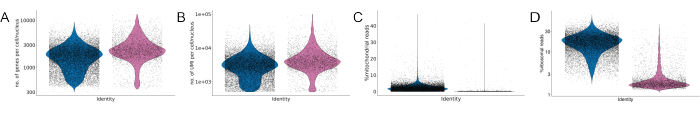
Figure 4: Quality control of scRNA- and snRNA-seq. Violin plots illustrate the absolute UMI count (A), gene count (B), mitochondrial (C), and ribosomal reads (D) of cells (blue) and nuclei (pink-purple). Please click here to view a larger version of this figure.
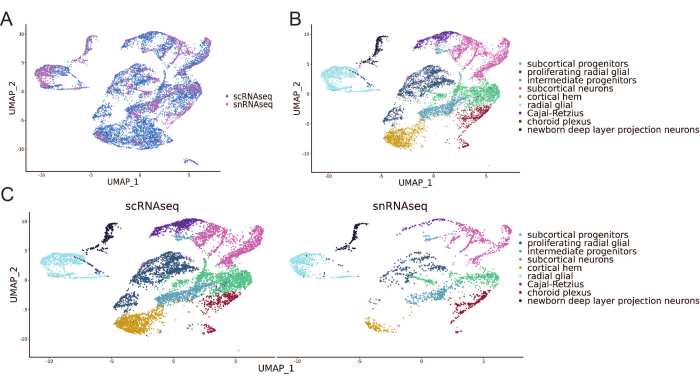
Figure 5: scRNA-seq and snRNA-seq retrieve similar cell populations in 30-day old organoids. Integrated embedded UMAP of 30-day old organoids, showing scRNA-seq (blue) and snRNA-seq results (pink-purple) (A), integrated and annotated and individual profiles of scRNA-seq and snRNA-seq (B,C). Please click here to view a larger version of this figure.
Table 1: Cell culture media and coating components Please click here to download this Table.
Table 2: Dissociation buffers for scRNA and snRNA-seq. Please click here to download this Table.
Supplementary Coding File 1: Coding file used to analyze the scRNA-seq and snRNA-seq data. Please click here to download this File.
Discussion
Transcriptomic analysis of single cells and single nuclei has emerged as a pivotal tool for understanding gene regulatory mechanisms within complex tissues. Both methods enable transcriptome studies of brain organoids. To ensure an overall successful experiment, the quality of the starting material is of high relevance. Therefore, it is necessary to cut the organoids regularly to prevent the formation of a necrotic core26. It is also possible to eliminate this issue with an Air-Liquid Interface culture27. To reduce batch effects due to organoid heterogeneity, we suggest using at least four organoids per extraction. Alternatively, it is possible to conduct multiple sequencing runs of individual organoids, which additionally allows examination of the variability within the same batch17. When using both methods, scRNA-seq and snRNA-seq, in parallel, cutting organoids into two and dividing them for each method can further reduce differences caused by organoid heterogeneity.
To obtain a good quality transcriptomic profile of an individual cell or nucleus, it is crucial that the cellular or nuclear membrane remains intact during the whole procedure. Therefore, it is crucial to optimize both the enzyme concentration and the timing of the enzymatic digestion, which need to be adjusted to the age and size of the organoid. Additionally, it is essential to use the correct temperatures and centrifugation speed and to avoid harsh pipetting. In case of low viability, a dead cell removal kit can be used after the dissociation of the organoids into single cells. Complementary, for the isolation of nuclei it is vital to carefully wash the organoid with PBS and adapt the number of strokes with the douncer to organoid size. It is also important to carefully layer the Percoll gradient in order to avoid disturbances in the layering. If damaged nuclei remain after isolation, it is advised to implement a sorting step via Fluorescence-activated cell sorting22. Overall, the use of RNAse inhibitors is vital for all steps from sample dissociation to library generation to preserve the high quality RNA of each cell or nucleus.
In this study, scRNA-seq yielded more cells, allowing for a broader overview of cell populations. Additionally, cells contain more RNA and are easier to enrich for specific cell types through surface markers as compared to nuclei. However, single cell dissociation relies on an enzymatic process which can induce the transcription of stress-related RNAs19,28. Especially in diseases that are associated with an increased stress response, this can result in dissociation-induced artifacts20. Moreover, it was shown that single cell isolation resulted in the loss of sensitive cell populations3,29. While this may not be evident in this study, it is a possible outcome when using older brain organoids which are characterized by a higher cellular diversity.
As both datasets retrieve the same cell populations, the choice of the isolation method strongly depends on research questions, tissue acquisition and time management. While after dissociation the fixation and storage of single cells from brain organoids in methanol is well established30, the isolation of single nuclei from frozen brain organoids still requires optimization.
Overall, our protocols provide a versatile and adaptable platform to investigate the transcriptomic profile of individual cells and nuclei in brain organoids, paving the way to in-depth investigations of developmental- and disease-related questions in the in vitro human brain models.
Offenlegungen
The authors have nothing to disclose.
Acknowledgements
We thank Valeria Fernandez-Vallone for the original instructions for the Miltenyi Neural Dissociation kit. We also thank the Genomics Technology Platform of the Max Delbrueck Centrum for providing the recipe for the NP40 lysis buffer and valuable advice setting up this protocol. We also thank Margareta Herzog and Alexandra Tschernycheff for the lab organizational support.
Materials
| 1,4-DITHIO-DL-THREIT-LSG., F. D. MOL.-BIOL., ~1 M IN H2O (DTT) | Sigma | 43816-10ML | |
| 1.5 ml DNA low binding tubes | VWR | 525-0130 | microcentrifuge tube |
| 10x Cellranger pipeline | analysis pipline | ||
| 15 ml Falcon | Falcon | Centrifuge tube | |
| 2-Mercaptoethanol (BME) | Life Technologies | 21985023 | |
| 50 ml Falcon | Falcon | Centrifuge tube | |
| A83-01 | Bio Technologies | 379762 | |
| Antibiotic/Antimycotic Solution (100X) | Life Technologies | 15240062 | |
| B-27 Plus Supplement | Life Technologies | 17504044 | |
| B-27 Supplement without vitamin A | Life Technologies | 12587010 | |
| Bovine serum albumin, fatty acid free (BSA) | Sigma Aldrich | A8806-5G | |
| cAMP | Biogems | 6099240 | |
| cAMP | Biogems | 6099240 | |
| C-CHIP NEUBAUER IMPROVED | VWR | DHC-N01 | |
| Cell strainer 40 µm | Neolab | 352340 | |
| Cell strainer 70 µm (white) Nylon | Sigma | CLS431751-50EA | |
| Chromium Controller & Next GEM Accessory Kit | 10X Genomics | 1000204 | |
| Chromium Next GEM Chip G Single Cell Kit, 16 rxns | 10X Genomics | 1000127 | |
| Chromium Next GEM Single Cell 3' Kit v3.1 | 10X Genomics | 1000268 | |
| Complete, EDTA-free Protease Inhibitor Cocktaill | Roche | 11873580001 | |
| DAPI | MERCK Chemicals | 0000001722 | |
| DMEM/F12 | Life Technologies | 11320074 | |
| Dounce tissue grinder set 2 mL complete | Sigma Aldrich | 10536355 | |
| Essential E8 Flex Medium | Life Technologies | A2858501 | |
| EVE Cell Counting Slides | VWR | EVS-050 ( 734-2676) | |
| Foetal bovine serum tetracycline free (FBS) | PAN Biotech | P30-3602 | |
| Geltrex LDEV-Free (coating) | Life Technologies | A1413302 | |
| gentleMACS | Miltenyi Biotec | dissociation maschine | |
| GlutaMAX supplements | Life Technologies | 35050038 | |
| Heparin sodium cell culture tested | Sigma | H3149-10KU | |
| human recombinant BDNF | StemCell Technologies | 78005.3 | |
| human recombinant GDNF | StemCell Technologies | 78058.3 | |
| Insulin Solution Human | Sigma Aldrich | I2643-25MG | |
| Knockout serum replacement | Life Technologies | 10828028 | |
| LDN193189 Hydrochloride 98% | Sigma Aldrich | 130-106-540 | |
| MEM non-essential amino acid (100x) | Sigma Aldrich | M7145-100ml | |
| MgCl2 Magnesium Chloride (1M) RNAse free | Thermo Scientific | AM9530G | |
| mTeSR Plus | StemCell Technologies | 100-0276 | stem cell medium |
| mTeSR1 | StemCell Technologies | 85850 | stem cell medium |
| N2 Supplement | StemCell Technologies | 17502048 | |
| Neural Tissue Dissociation Kit | Miltenyi Biotec B.V. & Co. KG | 130-092-628 | |
| Neurobasal Plus | Life Technologies | A3582901 | |
| NextSeq500 system | Illumina | Sequencer | |
| NP-40 Surfact-Amps Detergent Solution | Life Technologies | 28324 | |
| PBS Dulbecco’s | Invitrogen | 14190169 | |
| PenStrep (Penicillin – Streptomycin) | Life Technologies | 15140122 | |
| Percoll | Th. Geyer | 10668276 | |
| Pluronic (R) F-127 | Sigma Aldrich | P2443-1KG | |
| RiboLock RNase Inhibitor | Life Technologies | EO0382 | |
| Rock Inhibitor (Y-27632 dihydrochloride) SB | Biomol | Cay10005583-10 | |
| SB 431542 | Biogems | 3014193 | |
| Sodium chloride NaCl (5M), RNase-free-100 mL | Invitrogen | AM9760G | |
| StemFlex Medium | Thermo Scientific | A3349401 | stem cell medium |
| StemMACS iPS-Brew XF | Miltenyi Biotec | 130-104-368 | stem cell medium |
| TC-Platte 96 Well, round bottom | Sarstedt | 83.3925.500 | |
| TISSUi006-A | TissUse GmbH | https://hpscreg.eu/cell-line/TISSUi006-A | |
| Trypan Blue | T8154-20ml | Sigma | |
| TrypLE Express Enzyme, no phenol red | Life Technologies | 12604013 | Trypsin-based reagent |
| UltraPure 1M Tris-HCl Buffer, pH 7.5 | Life Technologies | 15567027 | |
| XAV939 | Enzo Life sciences | BML-WN100-0005 |
Referenzen
- Finkbeiner, S. R., et al. Stem cell-derived human intestinal organoids as an infection model for Rotaviruses. mBio. 3 (4), e00159-e00212 (2012).
- Freedman, B. S., et al. Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids. Nat Commun. 6, 8715 (2015).
- Guan, Y., et al. Human hepatic organoids for the analysis of human genetic diseases. JCI Insight. 2 (17), e94954 (2017).
- Lancaster, M. A., et al. Cerebral organoids model human brain development and microcephaly. Nature. 501 (7467), 373-379 (2013).
- Dang, J., et al. Zika virus depletes neural progenitors in human cerebral organoids through activation of the innate immune receptor TLR3. Cell Stem Cell. 19 (2), 258-265 (2016).
- Inak, G., et al. Defective metabolic programming impairs early neuronal morphogenesis in neural cultures and an organoid model of Leigh syndrome. Nat Commun. 12 (1), 1929 (2021).
- Karlsson, M., et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 7 (31), eabh2169 (2021).
- Piwecka, M., Rajewsky, N., Rybak-Wolf, A. Single-cell and spatial transcriptomics: deciphering brain complexity in health and disease. Nat Rev Neurol. 19 (6), 346-362 (2023).
- Lim, B., Lin, Y., Navin, N. Advancing cancer research and medicine with single-cell genomics. Cancer Cell. 37 (4), 456-470 (2020).
- Camp, J. G., et al. Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proc Natl Acad Sci U S A. 112 (51), 15672-15677 (2015).
- Fiorenzano, A., et al. Single-cell transcriptomics captures features of human midbrain development and dopamine neuron diversity in brain organoids. Nat Commun. 13 (1), 3312 (2022).
- Kanton, S., et al. Organoid single-cell genomic atlas uncovers human-specific features of brain development. Nature. 574 (7778), 418-422 (2019).
- Notaras, M., et al. Schizophrenia is defined by cell-specific neuropathology and multiple neurodevelopmental mechanisms in patient-derived cerebral organoids. Mol Psychiatry. 27 (3), 1416-1434 (2022).
- Rybak-Wolf, A., et al. Modelling viral encephalitis caused by herpes simplex virus 1 infection in cerebral organoids. Nat Microbiol. 8 (7), 1252-1266 (2023).
- Bock, C., et al. The organoid cell atlas. Nat Biotechnol. 39 (1), 13-17 (2021).
- Brazovskaja, A., Treutlein, B., Camp, J. G. High-throughput single-cell transcriptomics on organoids. Cur Opinion Biotechnol. 55, 167-171 (2019).
- Velasco, S., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 570, 523-527 (2019).
- Uzquiano, A., et al. Proper acquisition of cell class identity in organoids allows definition of fate specification programs of the human cerebral cortex. Cell. 185 (20), 3770-3788.e27 (2022).
- Mattei, D., et al. Enzymatic dissociation induces transcriptional and proteotype bias in brain cell populations. Int J Mol Sci. 21 (21), 7944 (2020).
- Van Den Brink, S. C., et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 14 (10), 935-936 (2017).
- Slyper, M., et al. A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med. 26 (5), 792-802 (2020).
- Santos, M. D., et al. Extraction and sequencing of single nuclei from murine skeletal muscles. STAR Protoc. 2 (3), 100694 (2021).
- Fleck, J. S., et al. Inferring and perturbing cell fate regulomes in human cerebral organoids. Nature. 621 (7978), 365-372 (2021).
- Martins-Costa, C., et al. Morphogenesis and development of human telencephalic organoids in the absence and presence of exogenous extracellular matrix. EMBO J. 42 (22), e113213 (2023).
- Hao, Y., et al. Integrated analysis of multimodal single-cell data. Cell. 184 (13), 3573-3587.e29 (2021).
- Choe, M. S., et al. A simple method to improve the quality and yield of human pluripotent stem cell-derived cerebral organoids. Heliyon. 7 (6), e07350 (2021).
- Giandomenico, S. L., et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 22 (4), 669-679 (2019).
- Denisenko, E., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 21 (1), 130 (2020).
- Wen, F., Tang, X., Xu, L., Qu, H. Comparison of single-nucleus and single-cell transcriptomes in hepatocellular carcinoma tissue. Mol Med Rep. 26 (5), 339 (2022).
- Alles, J., et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 15 (1), 44 (2017).

