A Viral Attachment Assay for Screening of Antiviral Compounds
Abstract
Source: Tai, C. et al., Early Viral Entry Assays for the Identification and Evaluation of Antiviral Compounds. J. Vis. Exp. (2015)
This video demonstrates a viral attachment assay for screening antiviral compounds. Using genetically modified hepatitis C virus and luciferase, it assesses inhibitory effects on viral attachment, as indicated by lower light intensity in co-treated wells compared to wells treated with the virus alone.
Protocol
NOTE: Ensure that all procedures involving cell culture and virus infection are conducted in certified biosafety hoods that are appropriate for the biosafety level of the samples being handled. For the purpose of describing the protocols, Gaussia luciferase reporter-tagged hepatitis C virus (HCV) is used as a model virus. In the context of the representative results, the compounds chebulagic acid (CHLA) and punicalagin (PUG) are used as candidate antivirals that target viral glycoprotein interactions with cell surface glycosaminoglycans during the early viral entry steps. Heparin, which is known to interfere with the entry of many viruses, is used as a positive control treatment in such a context. For basic background on virology techniques, propagation of viruses, determination of virus titer, and expression of infectious dose in plaque forming units (PFU), focus forming units (FFU), or multiplicity of infection (MOI), the reader is referred to reference. For prior examples and optimized conditions used for viruses shown in the representative results, the reader is referred to references as well as details listed in Table 1, Figure 1A.
1. Cell Culture, Compound Preparation, and Compound Cytotoxicity
- Grow the respective cell line for the virus infection system to be analyzed (Table 1). For HCV, grow the Huh-7.5 cells in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 200 U/ml penicillin G, 200 µg/ml streptomycin, and 0.5 µg/ml amphotericin B.
- Prepare the test compounds and controls using their respective solvents: for example, dissolve CHLA and PUG in dimethyl sulfoxide (DMSO); prepare heparin in sterile double-distilled water. For all subsequent dilutions, use culture media.
NOTE: The final concentration of DMSO in the test compound treatments is less than 1% in the experiments; 1% DMSO is included as a negative control treatment in the assays for comparison. - Determine the cytotoxicity of the test compounds (e.g., CHLA and PUG) on the cells for the viral infection by using cell viability determining reagent such as XTT (2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-5-phenylamino)-carbonyl]-2H-tetrazolium hydroxide):
- For HCV, seed Huh-7.5 cells in a 96-well plate (1 × 104 cells per well) and incubate at 37 °C in a 5% CO2 incubator O/N to obtain a monolayer.
- Apply DMSO control (1%) or increasing concentrations of the test compounds CHLA and PUG (ex. 0, 10, 50, 100, and 500 μM) to the culture wells in triplicate.
- Incubate at 37 °C for 72 hr, then discard the medium in the plate and wash the cells with 200 μl of phosphate-buffered saline (PBS) twice.
- Add 100 µl of assaying solution from the XTT-based in vitro toxicology assay kit to each well and incubate the plates at 37 °C for another 3 hr to allow XTT formazan production.
- Determine the absorbance with a microplate reader at a test wavelength of 492 nm and a reference wavelength of 690 nm.
- Calculate the percentage of surviving cells using the following formula: cell viability (%) = At / As × 100%, where 'At' and 'As' refer to the absorbance of the test compounds and the solvent control (ex. 1% DMSO) treatments, respectively. Determine the concentration of 50% cellular cytotoxicity (CC50) of the test compounds from an analytical software such as GraphPad Prism according to the manufacturer's protocol.
2. Readout of Viral Infection
NOTE: The readout of viral infection depends on the virus system used and can involve methods such as plaque assays or measuring reporter signals from reporter-tagged viruses. The method for detecting reporter-HCV infection based on the luciferase reporter activity is described below.
- Collect the supernatants from the infected wells and clarify at 17,000 x g in a microcentrifuge for 5 min at 4 °C.
- Mix 20 μl of test supernatant to 50 μl of luciferase substrate from the Gaussia luciferase assay kit and directly measure with a luminometer according to the manufacturer's instructions.
- Express HCV infectivity as log10 of relative light units (RLU) for determining viral inhibition (%) and calculate the 50% effective concentration (EC50) of the test compounds against HCV infection using algorithms from GraphPad Prism software according to the manufacturer's protocol.
3. Viral Attachment Assay
NOTE: Examples of incubation period and viral dose for various viruses are listed in Figure 1A, 'Attachment'. Higher concentrations of the virus can also be tested by increasing the MOI/PFU.
- Seed Huh-7.5 cells in a 96-well plate (1 × 104 cells per well) and incubate at 37 °C in a 5% CO2 incubator O/N to obtain a monolayer.
- Pre-chill the cell monolayers in plates at 4 °C for 1 hr.
- Co-treat the cells with HCV inoculum (MOI = 0.01) and test compounds or controls (final concentrations are: CHLA = 50 μM; PUG = 50 μM; heparin = 1,000 μg/ml; DMSO = 1%) at 4 °C for 3 hr. For example, to a 90 μl virus inoculum containing 1 x 102 FFU, add 10 μl of a 500 μM CHLA working dilution; this yields CHLA treatment at a final concentration of 50 μM and an infection by HCV at MOI = 0.01 on the cell monolayer.
NOTE: It is important to carry out the experiment at 4 °C since it allows for virus binding but precludes entry which most efficiently occurs at 37 °C. Perform the addition of virus and test compounds on ice and the ensuing incubation in a 4 °C refrigerator to ensure that the temperature is maintained at 4 °C. - Remove the supernatant and gently wash the cell monolayer with 200 μl of ice-cold PBS twice.
NOTE: Perform the washes gently to avoid lifting the cells. - Apply 100 μl of basal medium to each well and incubate at 37 °C for 72 hr.
- Analyze the resulting infection by assaying the supernatant for luciferase activity as described in '2. Readout of Viral Infection'.
Table 1: Host cell type for viral infection. The cell type used for each viral infection system described in the representative results is indicated. Additional details regarding the cells can be found in the reference.
| Virus | Cell Type |
| HCMV | HEL |
| HCV | Huh-7.5 |
| DENV-2 | Vero |
| MV | CHO-SLAM |
| RSV | HEp-2 |
Representative Results
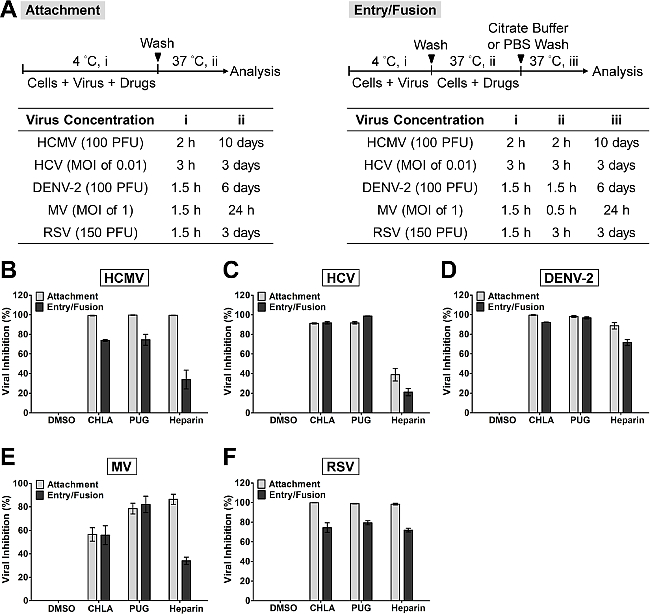
Figure 1. Evaluation of antiviral activities of the test compounds CHLA and PUG against virus attachment and entry/fusion. (A) The experimental procedure, virus concentration (PFU/well or MOI), and the time of addition and treatment with the test compounds (i, ii, iii) are presented for each virus in the schematics and the associated tables. In virus attachment analysis (light gray bars), monolayers of different cell types were pre-chilled at 4 °C for 1 hr, then co-treated with the respective viruses and test compounds at 4 °C (1.5 – 3 hr; i) before washing off the inoculates and test compounds for subsequent incubation (37 °C; ii) and examination of virus infection. In virus entry/fusion analysis (dark gray bars), seeded cell monolayers were pre-chilled at 4 °C for 1 hr and then challenged with the respective viruses at 4 °C for 1.5 – 3 hr (i). Cells were then washed and treated with the test compounds for an additional incubation period (ii) during which the temperature was shifted to 37 °C to facilitate the viral entry/fusion event. At the end of the incubation, extracellular viruses were removed by either citrate buffer (pH 3.0) or PBS washes, and the cells were further incubated (iii) for analysis of virus infection. Results for (B) HCMV, (C) HCV, (D) DENV-2, (E) MV, and (F) RSV are indicated in each additional panel. Data are plotted against the DMSO negative control treatment of virus infection and are presented as means ± SEM from three independent experiments.
Divulgaciones
The authors have nothing to disclose.
Materials
| DMEM | GIBCO | 11995-040 | |
| FBS | GIBCO | 26140-079 | |
| Penicillin-Streptomycin | GIBCO | 15070-063 | |
| Amphotericin B | GIBCO | 15290-018 | |
| DMSO | Sigma | D5879 | |
| In vitro toxicology assay kit, XTT-based | Sigma | TOX2 | |
| PBS pH 7.4 | GIBCO | 10010023 | |
| Microplate reader | Bio-Tek Instrument, Inc. | ELx800 | |
| Microcentrifuge | Thermo Scientific | 75002420 | |
| BioLux Gaussia luciferase assay kit | New England Biolabs | E3300L | |
| Luminometer | Promega | GloMax-20/20 |

