Analysis of Cap-binding Proteins in Human Cells Exposed to Physiological Oxygen Conditions
Summary
Here, we present human cell culture protocols to analyze translation initiation factors that bind the 5′ cap of mRNA during physiological oxygen conditions. This method utilizes an Agarose-linked m7GTP cap analog and is suitable to investigate cap-binding factors and their interacting partners.
Abstract
Translational control is a focal point of gene regulation, especially during periods of cellular stress. Cap-dependent translation via the eIF4F complex is by far the most common pathway to initiate protein synthesis in eukaryotic cells, but stress-specific variations of this complex are now emerging. Purifying cap-binding proteins with an affinity resin composed of Agarose-linked m7GTP (a 5′ mRNA cap analog) is a useful tool to identify factors involved in the regulation of translation initiation. Hypoxia (low oxygen) is a cellular stress encountered during fetal development and tumor progression, and is highly dependent on translation regulation. Furthermore, it was recently reported that human adult organs have a lower oxygen content (physioxia 1-9% oxygen) that is closer to hypoxia than the ambient air where cells are routinely cultured. With the ongoing characterization of a hypoxic eIF4F complex (eIF4FH), there is increasing interest in understanding oxygen-dependent translation initiation through the 5′ mRNA cap. We have recently developed a human cell culture method to analyze cap-binding proteins that are regulated by oxygen availability. This protocol emphasizes that cell culture and lysis be performed in a hypoxia workstation to eliminate exposure to oxygen. Cells must be incubated for at least 24 hr for the liquid media to equilibrate with the atmosphere within the workstation. To avoid this limitation, pre-conditioned media (de-oxygenated) can be added to cells if shorter time points are required. Certain cap-binding proteins require interactions with a second base or can hydrolyze the m7GTP, therefore some cap interactors may be missed in the purification process. Agarose-linked to enzymatically resistant cap analogs may be substituted in this protocol. This method allows the user to identify novel oxygen-regulated translation factors involved in cap-dependent translation.
Introduction
Translational control is emerging as an equally important step to transcriptional regulation in gene expression, especially during periods of cellular stress1. A focal point of translation control is at the rate-limiting step of initiation where the first steps of protein synthesis involve the binding of the eukaryotic initiation factor 4E (eIF4E) to the 7-methylguanosine (m7GTP) 5' cap of mRNAs2. eIF4E is part of a trimeric complex named eIF4F that includes eIF4A, an RNA helicase, and eIF4G, a scaffolding protein required for the recruitment of other translation factors and the 40S ribosome3. Under normal physiological conditions, the vast majority of mRNAs are translated via a cap-dependent mechanism, but under periods of cellular stress approximately 10% of human mRNAs contain 5' UTRs that could allow cap-independent translation intiation1,4. Cap-dependent translation has been historically synonymous with eIF4F, however, stress-specific variations of eIF4F have become a trending topic5-8.
Various cellular stresses cause eIF4E activity to be repressed via the mammalian target of rapamycin complex 1 (mTORC1). This kinase becomes impaired under stress, which results in the increased activity of one of its targets, the 4E-binding protein (4E-BP). Non-phosphorylated 4E-BP binds to eIF4E and blocks its ability to interact with eIF4G causing the repression of cap-dependent translation9,10. Interestingly, a homolog of eIF4E named eIF4E2 (or 4EHP) has a much lower affinity for 4E-BP11, perhaps allowing it to evade stress-mediated repression. Indeed, initially characterized as a repressor of translation due to its lack of interaction with eIF4G12, eIF4E2 initiates the translation of hundreds of mRNAs that contain RNA hypoxia response elements in their 3' UTR during hypoxic stress6,13. This activation is achieved through interactions with eIF4G3, RNA binding protein motif 4, and the hypoxia inducible factor (HIF) 2α to constitute a hypoxic eIF4F complex, or eIF4FH6,13. As a repressor under normal conditions, eIF4E2 binds with GIGYF2 and ZNF59814. These complexes were, in part, identified through Agarose-linked m7GTP affinity resins. This classic method15 is standard in the field of translation and is the best and most commonly used technique to isolate cap-binding complexes in pull down and in vitro binding assays16-19. As the cap-dependent translation machinery is emerging as flexible and adaptable with inter-changing parts6-8,13, this method is a powerful tool to rapidly identify novel cap-binding proteins involved in the stress response. Furthermore, variations in eIF4F could have broad implications as several eukaryotic model systems appear to use an eIF4E2 homolog for stress responses such as A. thaliana20, S. Pombe21, D. melanogaster22, and C. elegans23.
Evidence suggests that variations in eIF4F may not be strictly limited to stress conditions, but be involved in normal physiology24. The oxygen supply to tissues (at capillary ends) or within tissues (measured via microelectrodes) varies from 2-6% in the brain25, 3-12% in the lungs26, 3.5-6% in the intestine27, 4% in the liver28, 7-12% in the kidney29, 4% in muscle30, and 6-7% in bone marrow31. Cells and mitochondria contain less than 1.3% oxygen32. These values are much closer to hypoxia than the ambient air where cells are routinely cultured. This suggests that what were previously thought of as hypoxia-specific cellular processes may be relevant in a physiological setting. Interestingly, eIF4F and eIF4FH actively participate in the translation initiation of distinct pools or classes of mRNAs in several different human cell lines exposed to physiological oxygen or "physioxia"24. Low oxygen also drives proper fetal development33 and cells generally have higher proliferation rates, longer lifespans, less DNA damage and less general stress responses in physioxia34. Therefore, eIF4FH is likely a key factor in the expression of select genes under physiological conditions.
Here, we provide a protocol to culture cells in fixed physiological oxygen conditions or in a dynamic fluctuating range that is likely more representative of tissue microenvironments. One advantage of this method is that cells are lysed within the hypoxia workstation. It is not often clear how the transition from hypoxic cell culture to cell lysis is performed in other protocols. Cells are often first removed from a small hypoxia incubator before lysis, but this exposure to oxygen could affect biochemical pathways as the cellular response to oxygen is rapid (one or two min)35. Certain cap-binding proteins require interactions with a second base or can hydrolyze the m7GTP, therefore some cap interactors may be missed in the purification process. Agarose-linked to enzymatically resistant cap analogs may be substituted in this protocol. Exploring the activity and composition of eIF4FH and other variations of eIF4F through the method described here will shed light on the intricate gene expression machineries that cells utilize during physiological conditions or stress responses.
Protocol
1. Preparations for Cell Culture
- Purchase commercially available stocks of human cells.
NOTE: This protocol utilizes HCT116 colorectal carcinoma and primary human renal proximal tubular epithelial cells (HRPTEC). - Make 500 ml medium for culture of HCT116: Dulbecco's Modified Eagle Medium (DMEM)/High glucose medium supplemented with 7.5% Fetal Bovine Serum (FBS) and 1% Penicillin/Streptomycin (P/S).
- Make 500 ml medium for culture of HRPTEC: Epithelial cell medium supplemented with 5% FBS, 1% epithelial cell growth supplement, and 1% P/S.
- Prepare 500 ml of 1x Phosphate-Buffered Saline (PBS): 140 mM NaCl, 3 mM KCl, 10 mM Na2HPO4, 15 mM KH2PO4. Adjust pH to 7.4 and sterilize by autoclaving for 40 min at 121 °C.
2. Initiation of Cell Culturing
- Carefully aspirate the medium from a 80-90% confluent dish without disturbing the cells.
- Aliquot 3-5 ml of 1x PBS per culture dish, gently rock each flask to coat the surface area of the dish, and carefully aspirate the 1x PBS. Repeat for a second 1x PBS wash.
- Aliquot 3 ml of 0.05% trypsin-ethylenediaminetetraacetic acid (EDTA) into each culture dish and gently rock to evenly coat the cells with trypsin-EDTA. Incubate at 37 °C for 2-3 min.
- Transfer the detached cells into a 15 ml centrifuge tube and centrifuge at 4,000 x g for 90 sec.
- Resuspend cell pellet in 1 ml of complete DMEM.
- Count cells using a haemocytometer. Calculate how many non-pyrogenic and polystyrene 100 mm cell culture dishes are required to achieve an initial seeding density of approximately 2,500-5,000 cells per cm2.
- Pre-warm complete cell culture medium made in steps 1.2 or 1.3 in a 37 °C water bath for 30-45 min.
- Decontaminate the medium bottle and cell vial with 70% ethanol. From this point forward all operations should be performed aseptically.
- Aliquot 6 ml of complete DMEM into as many 100 mm cell culture dishes required to use the entire volume of frozen cells within the seeding density mentioned in step 2.6.
- Transfer the volume of cell suspension calculated in step 2.6 to each of the 100 mm culture dishes. Rock each plate manually to evenly distribute the cells over the surface of the plate.
- Place the seeded 100 mm cell culture dish in the incubator at 37 °C in a 5% CO2 atmosphere. Allow cells to incubate at least 24 hr before the next steps.
3. Subculturing
- View each cell culture dish under a microscope to determine the percentage of cell confluency (% cells covering the area of the dish). Return cells to the incubator and pre-warm complete medium and 1x PBS. Proceed to the next step if the cells are 80-100% confluent.
- Carefully aspirate the spent medium without disturbing the cells in each culture dish.
- Aliquot 3-5 ml of 1x PBS per culture dish, gently rock each flask to coat the surface area of the dish, and carefully aspirate the 1x PBS. Repeat for a second 1x PBS wash.
- Aliquot 3 ml of 0.05% trypsin-EDTA into each culture dish and gently rock to evenly coat the cells with trypsin-EDTA. Incubate at 37 °C for 2-3 min.
- During this incubation, aliquot 10 ml of complete medium into as many culture dishes are required to obtain cell density of 2,500-5,000 cells per cm2.
- When the cells have detached from the plate, quickly add 3 ml of 1x defined trypsin inhibitor and gently swirl the dish for 1 min to ensure that all of the trypsin-EDTA has been neutralized.
- Transfer the detached cells into a sterile 15 ml centrifuge tube and set aside. Add 3 ml 1x PBS to the dish to gather any remaining cells, and transfer that to the same 15 ml centrifuge tube.
- Pellet the cells by centrifuging 150 x g for 5 min.
- Aspirate the supernatant and resuspend the cell pellet in 3 ml of complete medium.
- Count cells using a haemocytometer and seed 150 mm culture dishes containing 15 ml of complete medium to obtain a cell density of 2,500-5,000 cells per cm2. Use two 150 mm for each cap-binding assay.
NOTE: Oxygen diffusion to cells through liquid media is dependent on the volume36. It is recommended to keep the volume of the media consistent between experiments whether adherent cells or cells in suspension are used. Media can be pre-conditioned (incubated in the workstation for 24 hr as discussed in Discussion) to avoid these variabilities in diffusion. - Place the seeded-culture dishes in the incubator at 37 °C in a 5% CO2 atmosphere. Allow cells to incubate at least 24 hr before proceeding to the next steps.
4. Physioxic Exposure
- View the cells under the microscope. For best results, ensure that cells are 70-80% confluent for a 24 hr exposure, 50-60% confluent for a 48 hr exposure, and 40-50% confluent for a 72 hr exposure.
- Place the cells into a hypoxia workstation for the desired time (24, 48, or 72 hr depending on the experiment).
- Set the workstation to the appropriate physioxia.
NOTE: For example, a 3% O2 setting would be accompanied by 5% CO2 and 92% N2.
NOTE: If oxygen fluctuations within a dynamic range are desired over a specific timeline, program the schedule into the instrument or manually adjust the oxygen settings at the desired intervals. - Set the humidity to 60%. The workstation atmosphere is very dry nonetheless, and media will noticeably evaporate during long experiments (>48 hr). Keep a media reserve in a cell culture flask within the workstation (so that the media is equilibrated with the workstation atmosphere) to replenish the media in the cell culture dishes.
NOTE: Any solution that will interact with the cells prior to lysis (such as PBS and trypsin-EDTA) should be pre-conditioned for 24 hr before use within the hypoxia workstation so that the dissolved oxygen equilibrates with the atmospheric oxygen36. - Keep cells within the workstation until lysis.
5. Preparing Buffers for Cap-binding Assay
- Prepare 1x Tris-buffered Saline (TBS).
- In 800 ml of dH2O, dissolve 8.76 g NaCl and 6.05 g Tris Base.
- Bring the solution to a final pH of 7.4 using 1 M HCl.
- Bring the final volume to 1 L using dH2O.
- Prepare non-denaturing lysis buffer. Prepare lysis buffer the day of the cap-binding assay. Make extra lysis buffer to wash the blank agarose beads and the γ-aminophenyl-m7GTP agarose C10-linked beads (steps 7.9 and 7.13).
- In 7 ml of dH2O, add 160 µl 5 M NaCl, 160 µl 1 M Tris-HCl pH 7.4, 40 µl 200 mM NaF, 40 µl 1 M MgCl2, and 40 µl 1 mM sodium orthovanadate.
- Add 40 µl of Igepal to the 7 ml solution. Cut the tip of the pipette to help retrieve Igepal as it is extremely viscous.
- Mix by pipetting up and down, or vortex, the 7 ml solution until all of the Igepal has been dissolved.
- Raise the final volume of the solution to 8 ml and keep on ice.
- Prepare 4x Sodium Dodecyl Sulfate (SDS)-PAGE Sample Buffer.
- In a beaker, combine 16 ml 1 M Tris-HCl pH 6.8, 12.64 ml glycerol, and 8 ml dithiothreitol (DTT).
- Add 3.2 g of SDS and 0.16 g of bromophenol blue. Allow powder to fully dissolve by mixing with a magnetic stir bar.
- Aliquot into 1.5 ml microcentrifuge tubes and store at -20 °C.
6. Cell Lysis
- Prepare non-denaturing lysis buffer and keep on ice the day of the cap-binding assay.
- Pre-warm 1x PBS and trypsin in a 37 °C water bath for 30 min.
- Aliquot 980 µl of lysis buffer into a 1.5 ml microcentrifuge tube for each cap-binding assay being performed.
- Add 10 µl of 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride and 10 µl of 100x protease inhibitor cocktail to the 980 µl of lysis buffer. Keep on ice.
- Discard the media from each 150 mm dish in a waste container within the hypoxia workstation.
- Wash the cells with 3-4 ml of warm 1x PBS and discard all of the liquid.
- Add 1 ml of warm 0.05% trypsin-EDTA to each dish and let sit for 2 min or until cells are no longer adhered to the plate.
- Using a pipette, transfer the cells of one plate to a 1.5 ml microcentrifuge tube. Repeat as needed so that each plate of cells is transferred to a separate 1.5 ml microcentrifuge tube.
- Centrifuge the 1.5 ml microcentrifuge tubes at 6,000 x g for 90 sec to pellet the cells.
- Aspirate the trypsin using a pipette without disturbing the pellet. If the pellet is disturbed, re-centrifuge the 1.5 ml microcentrifuge tube at 6,000 x g for 90 sec.
- Wash the cells with warm 1x PBS by gently pipetting 200 µl of PBS into the tube just above the pellet. Re-centrifuge if the pellet is disturbed.
- Aspirate the PBS without disturbing the pellet.
- Pipette 500 µl from the 1 ml prepared lysis buffer solution onto the first cellular pellet. Resuspend the pellet entirely by pipetting up and down.
- Combine the resuspended cells with the second cell pellet and resuspend the second pellet by pipetting up and down. Repeat this process until all pellets have been combined and resuspended for each sample.
- Combine the lysate with the remaining 500 µl of lysis buffer in a 1.5 ml microcentrifuge tube.
- Remove samples from the hypoxia workstation.
NOTE: After adding lysis buffer, all subsequent steps are to be performed in ambient air as the hypoxia workstation is set to 37 °C and the following steps are to be performed cold (4 °C or on ice). - Lyse the cells using gentle agitation by rotating the samples at 4 °C for 1.5-2 hr.
- Centrifuge the lysed cells at 12,000 x g for 15 min at 4 °C to remove cellular debris.
- Transfer the lysate to a new 1.5 ml microcentrifuge tube and discard the pellet.
- Reserve a portion (5-10%) of the supernatant to be used as a whole cell lysate input control for future western blot analysis.
7. Cap-binding Assay
- For each sample, transfer 50 µl of the blank agarose bead control slurry and 50 µl of the γ-aminophenyl-m7GTP agarose C10-linked bead slurry to separate 1.5 ml microcentrifuge tubes. Use scissors to remove the tip of the pipette to facilitate collection of the bead slurry.
- Pellet the beads by centrifuging the slurry at 500 x g for 30 sec.
- Remove the supernatant carefully and resuspend the beads in 500 µl of TBS.
- Repeat steps 7.2 and 7.3. Pellet the beads at 500 x g for 30 sec and remove the supernatant.
- Transfer the supernatant containing the lysate from step 6.19 to the 1.5 ml microcentrifuge tube containing the blank agarose beads.
NOTE: The blank agarose beads act as a pre-clearing step to remove proteins from the lysate that non-specifically interact with the bead. - Incubate for 10 min at 4 °C with gentle agitation.
- Pellet the blank agarose beads by centrifuging at 500 x g for 30 sec.
- Transfer the lysate to the 1.5 ml microcentrifuge tube containing the γ-aminophenyl-m7GTP agarose C10-linked beads.
- Wash the blank agarose beads by resuspending the beads in 500 µl of lysis buffer, pelleting the beads by centrifuging at 500 x g for 30 sec, and discarding the supernatant. Repeat this four times for a total of five washes.
- Resuspend the blank agarose beads in 1x SDS-PAGE sample buffer, and boil the beads for 90 sec at 95 °C. Store at -20 °C for future western blot analysis to observe whether the protein of interest binds non-specifically to the bead.
- Incubate the lysate with the γ-aminophenyl-m7GTP agarose C10-linked beads with gentle agitation for 1 hr at 4 °C to capture cap-binding proteins.
- Pellet the γ-aminophenyl-m7GTP agarose C10-linked beads by centrifuging at 500 x g for 30 sec. Discard the supernatant.
- Wash the γ-aminophenyl-m7GTP agarose C10-linked beads by repeating step 7.9.
- Resuspend the beads in 600 µl of lysis buffer.
- Add GTP to a final concentration of 1 mM.
- Incubate the γ-aminophenyl-m7GTP agarose C10-linked beads + 1 mM GTP with gentle agitation for 1 hr at 4 °C. Note: This will disassociate proteins that non-specifically interact with m7GTP (i.e., also interact with non-methylated GTP).
- Pellet the γ-aminophenyl-m7GTP agarose C10-linked beads by centrifuging at 500 x g for 30 sec.
- Transfer the supernatant to a new 1.5 ml microcentrifuge tube and add 200 µl of 4x SDS-PAGE sample buffer (50 mM Tris-HCl, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol). Store the GTP control sample at -20 °C for future western blot analysis37. Wash the beads by repeating step 7.9.
- Resuspend the beads in 1x SDS-PAGE sample buffer (50 mM Tris-HCl, 100 mM DTT, 2% SDS, 0.1% bromophenol blue, and 10% glycerol), and boil at 95 °C for 90 sec. Store the m7GTP-bound fraction at -20 °C for future western blot analysis37.
Representative Results
Analysis of Cap-binding Ability in Response to Oxygen of eIF4E and eIF4E2 in an m7GTP Affinity Column
Figures 1 and 2 represent western blots of typical m7GTP affinity purification of two major cap-binding proteins in response to oxygen fluctuations in two human cell lines: primary human renal proximal tubular epithelial cells (HRPTEC) in Figure 1 and colorectal carcinoma (HCT116) in Figure 2. Cells are maintained at the indicated oxygen availability for 24 hr to ensure that the dissolved oxygen in the liquid media has equilibrated with the air within the hypoxia workstation. The input lane (IN) represents 10% of the whole cell lysate before the m7GTP-linked agarose beads are added to capture cap-binding proteins. This input lane is used as a baseline to measure enrichment of a target protein in the m7GTP pulldown. The GTP lane is the eluate from the m7GTP beads that were elute with 1 mM GTP. This step allows for the detection of proteins that also recognize non-methylated GTP. The cap-binding proteins eIF4E and eIF4E2 recognize m7GTP specifically, but their homolog eIF4E3 (not shown here) also binds to non-methylated GTP. The ability of eIF4E and eIF4E2 to bind the 5' mRNA cap structure (m7GTP) can be measured by comparing the intensity of their bands in the m7GTP column relative to the intensity in the input lane (total available pool).
This quantification can be performed by measuring the pixel intensity of the protein bands for three biological replicates with an image analysis software such as ImageJ. Results are expressed as relative means of density units (RDU) ± standard error of the mean. Raw values prior to normalization from both experimental and control samples were compared by unpaired two-tailed Student's t-test. P < 0.05 was considered statistically significant. This representative experiment shows that the two cell lines have different ranges of oxygen for their eIF4E and eIF4E2 usage. Figure 1 shows that in HRPTEC only eIF4E binds strongly to m7GTP at 8% O2 (Figure 1A), that both eIF4E and eIF4E2 are significantly associated with m7GTP from 3-5% O2 (Figure 1B–C), and that only eIF4E2 binds strongly to m7GTP at 1% O2 (Figure 1D). In Figure 2, in HCT116 cells, the oxygen-dependent usage of these cap-binding proteins is different: only eIF4E binds significantly to m7GTP at 12% O2 (Figure 2A), both cap-binding proteins bind significantly to m7GTP in the 5-8% O2 range (Figure 2B–C), and only eIF4E2 binds significantly to m7GTP at 3% O2 (Figure 2D). The absence of elution from the m7GTP column with GTP shows that eIF4E and eIF4E2 are specific to m7GTP and do not recognize non-methylated GTP. The bar graphs represent quantification of three biological replicates using ImageJ. Our results demonstrate that two major cap-binding proteins, eIF4E and eIF4E2, differ in their ability to bind the mRNA m7GTP cap structure in an oxygen-dependent manner.
Based on previous literature that these two proteins initiate the translation of unique classes of mRNAs, this shift in cap-binding activity could result in different proteomes generated to adapt to changes in oxygen availability. This technique can be used to characterize novel translation initiation factors that interact with the 5' mRNA cap (or with cap-binding proteins) through a targeted western blot approach or a broad approach such as mass spectrometry analysis of an m7GTP eluate.
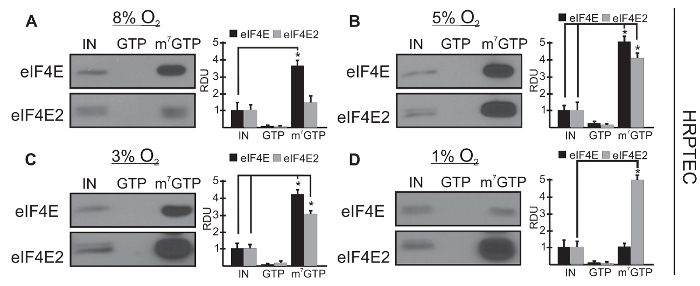
Figure 1: eIF4E and eIF4E2 activities overlap during physioxia in HRPTEC from 3-5% O2. Capture assays using m7GTP beads in human renal proximal tubular epithelial cell (HRPTEC) lysates exposed to (A) 8%, (B) 5%, (C) 3% or (D) 1% O2 for 24 hr. GTP, GTP wash to measure specificity for m7GTP; m7GTP, proteins bound to m7GTP beads after GTP wash. Representative Western blots are shown and data from at least three independent experiments were quantified by ImageJ and expressed as relative density units (RDU) relative to 10% input (IN) of whole cell lysate. *P < 0.05 (unpaired two-tailed Student's t-test) was considered a significant change when comparing the enrichment of eIF4E or eIF4E2 in the cap-bound fraction (m7GTP) to the IN. Data, mean ± standard error of the mean (SEM) of three independent experiments. This research was originally published in the Journal of Biological Chemistry. Timpano, S. and Uniacke, J. Human cells cultured under physiological oxygen utilize two cap-binding proteins to recruit distinct mRNAs for translation. 2016; 291(20): 10772-8224. Please click here to view a larger version of this figure.
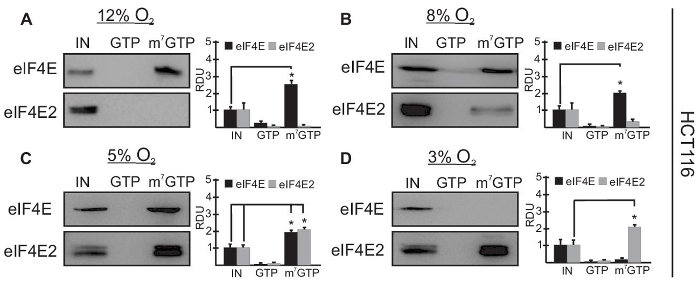
Figure 2. eIF4E and eIF4E2 activities overlap during physioxia in HCT116 from 5-8% O2. Capture assays using m7GTP beads in HCT116 human colorectal carcinoma lysates exposed to (A) 12%, (B) 8%, (C) 5% or (D) 3% O2 for 24 hr. GTP, GTP wash to measure specificity for m7GTP; m7GTP, proteins bound to m7GTP beads after GTP wash. Representative Western blots are shown and data from at least three independent experiments were quantified by ImageJ and expressed as relative density units (RDU) relative to 10% input (IN) of whole cell lysate. *P < 0.05 (unpaired two-tailed Student's t-test) was considered a significant change when comparing the enrichment of eIF4E or eIF4E2 in the cap-bound fraction (m7GTP) to the IN. Data, mean ± standard error of the mean (SEM) of three independent experiments. This research was originally published in the Journal of Biological Chemistry. Timpano, S. and Uniacke, J. Human cells cultured under physiological oxygen utilize two cap-binding proteins to recruit distinct mRNAs for translation. 2016; 291(20): 10772-8224. Please click here to view a larger version of this figure.
Discussion
The analysis of cap-binding proteins in human cells exposed to physiological oxygen conditions can allow for the identification of novel oxygen-regulated translation initiation factors. The affinity of these factors for the 5' cap of mRNA or other cap-associated proteins can be measured by the strength of their association to m7GTP-linked Agarose beads. One caveat of this technique is that it measures the cap-binding potential of proteins post-lysis, but it is performed under non-denaturing conditions that maintain protein-protein interactions and post-translational modifications (PTMs). Several protein-protein interactions mediate the binding of the eIF4E family to the 5' cap. The 4E-BP binds and represses eIF4E by blocking its interaction with eIF4G and preventing the recruitment of the 43S pre-initiation complex38. The eIF4E/4E-BP complex remains bound to the 5' mRNA cap, blocking any initiation from that transcript, and can be used as a marker for eIF4E inhibition in m7GTP affinity columns. Conversely, the interaction of eIF4E with eIF4G on m7GTP columns could be a marker for active translation initiation. PTMs are another method to regulate the activity of translation initiation factors. For example, phosphorylation of eIF4E by Mnk1 can increase its affinity for the 5' mRNA cap39. The protein modifier encoded by the Interferon Stimulated Gene 15 (ISG15) is a PTM that increases the cap-binding activity of eIF4E2 in response to interferon induction via pathogen infection40. The ISG15 gene contains a hypoxia response element in its promoter suggesting that this system could regulate eIF4E2 in low oxygen. Very little is known about how PTMs may alter the activity of cap-binding proteins during physiologically relevant oxygen conditions. This technique followed by mass spectrometry analysis could reveal novel oxygen-regulated and physiologically relevant PTMs that mediate the activity of translation initiation factors.
An alternative method to capturing with an m7GTP cap analog would be to immunoprecipitate a known cap-binding protein such as eIF4E or eIF4G and perform mass spectrometry to identify associated proteins. A limitation of this strategy is that cap-binding proteins often have alternative cellular roles such as within stress granules41 or in cap-independent translation1,4. Therefore, utilizing m7GTP cap analogs is the best, most reliable method to isolate cap-binding proteins, especially when investigating novel cap-associated proteins. A limitation of utilizing an m7GTP cap analog is that some cap-binding proteins such as the scavenger decapping enzyme DcpS not only interact with the m7GTP mRNA cap, but also require the second base for binding42. Furthermore, decapping enzymes in general, such as the Dcp1/Dcp2 complex, can hydrolyze m7GTP and effectively reduce resin capacity. Therefore, some cap-binding proteins may be lost in this analysis. To bypass these caveats, enzymatically resistant mRNA cap analogs can be utilized. Two non-hydrolyzable cap analogs, mononucleotide m7GpCH2pp or dinucleotide m7GpCH2ppA have been shown to capture DcpS, Dcp1, and Dcp243. These mRNA cap analogs are not available commercially, but must be chemically synthesized de novo. Another limitation of this protocol is that only cap-binding proteins or proteins interacting with cap-binding proteins via "piggybacking" will be captured. While cap-dependent translation initiation is the major form of initiation, cap-independent mechanisms are especially prevalent during conditions of cellular stress1. However, in the context of this technique and emerging trends in the field of translation initiation6-8,24, identifying novel stress-induced factors that participate in cap-dependent initiation is a promising area of research.
Another factor to consider in this protocol is oxygen in the media. Culturing cells in a liquid medium provides a barrier between the cells and the atmosphere. It has been shown that liquid media can take up to 24 hr to equilibrate with the atmospheric gas composition36. Therefore, cells must be cultured for 24 hr in the desired oxygen availability prior to lysis, or the media can be pre-conditioned if shorter incubations are required. Pre-conditioning involves maintaining the media in the hypoxia workstation for at least 24 hr in a sterile culture flask that allows gas exchange. Any oxygenated solution introduced to the cells within the hypoxia workstation could rapidly alter cellular signaling pathways such as the cap-dependent translation initiation that is being examined in this protocol. Due to the sensitivity of the oxygen-sensing pathways in cells, any solution that will interact with cells prior to lysis such as PBS and trypsin-EDTA must also be pre-conditioned for at least 24 hr. Lysis and post-lysis wash buffers must be used cold and are therefore not pre-conditioned in the 37 °C hypoxia workstation. While some oxygen-dependent modifications to proteins could be active post-lysis, the cold buffers are required to minimize enzymatic activities and "shuffling" of protein-protein or protein-RNA complexes that could lead to artefacts. A modification to this protocol to increase stringency may be to pre-condition lysis and post-lysis wash buffers for 24 hr and then incubate them on ice within the workstation before use. Media and buffers should not be kept in the hypoxia workstation for more than three days because these solutions will concentrate due to water evaporation. For longer term studies (>3 days), the initial amount of cells seeded must be carefully considered. As cells divide and the surface area of the dish becomes crowded, other stresses such as media acidification could affect the activity of cap-binding proteins. On the final day of the experiment, cells should have a confluency of 80-90%.
There are four critical steps within this protocol: maintaining the cells in the hypoxia workstation until lysis, the amount of cells used, washing the beads, and the non-methylated GTP control. 1) There are several methods to maintain cells in low oxygen. Most of these include small chambers that can hold only cell culture dishes, but any manipulation or cell lysis must be performed outside the chambers in ambient air. Components of the oxygen-sensing machinery such as the hypoxia inducible factors are activated or inactivated in a matter of one or two minutes35. Therefore, as mentioned previously, lysing the cells in a hypoxia workstation is critical to ensure the activity of the cap-binding proteins is associated with the respective oxygen availability. 2) The number of cells suggested in this protocol (two 150 mm dishes) is adequate for strong m7GTP interactors such as the eIF4E family or members of the eIF4F complex (eIF4A and eIF4G). More cells, at least five 150 mm dishes, could be required to detect proteins with transient interactions or that only associate with the m7GTP mRNA cap through eIF4F. 3) Washing the beads after incubating them with the cell lysate can be performed in a stringent and less stringent manner. The protocol presented here offers a stringent option where lysis buffer is used to wash the beads five times. If proteins with weaker interactions to cap-binding proteins are of interest, one can perform fewer washes and utilize Tris-buffered saline instead of lysis buffer. 4) While it is important to pre-clear the cell lysate with unconjugated agarose to remove proteins that non-specifically interact with the bead, some cap-binding proteins cannot differentiate between the true mRNA cap structure, methylated GTP (m7GTP), and non-methylated GTP. One such protein is the eIF4E homologue eIF4E344. Eluting the beads with GTP will dislodge any protein that also recognizes non-methylated GTP, which could be of interest. The biological relevance of proteins that recognize both m7GTP and GTP has yet to be fully elucidated. This is emphasized by the few publications involving eIF4E3 (which is a bona fide cap-binding protein with a conserved cap-binding domain) relative to the eIF4E and eIF4E2 proteins, which do recognize m7GTP from non-methylated GTP.
This technique, as presented, can be performed as a targeted approach to identify m7GTP interactors of interest or as a broad approach by analyzing the m7GTP eluate via mass spectrometry. Several other techniques can follow physioxic exposure (protocol 4) to analyze cap-binding activity such as subcellular fractionation, immunofluorescence, co-immunoprecipitation, and polysome analysis. Translational control is emerging as an equal contributor to gene expression as transcription. Utilizing this technique to investigate the activity and composition of cap-binding complexes will shed light on how cap-dependent translation initiation is regulated in response to low oxygen.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Natural Sciences and Engineering Council of Canada and the Ontario Ministry of Research and Innovation.
Materials
| γ-aminophenyl-m7GTP agarose C10-linked beads | Jena Bioscience | AC-1555 | Agarose-linked m7GTP |
| 100 mm culture dish | Corning | 877222 | 10-cm culture dish |
| 150 mm culture dish | Thermofisher | 130183 | 15-cm culture dish |
| AEBSF Hydrochloride | ACROS Organics | A0356829 | AEBSF |
| Agarose Beads | Jena Bioscience | AC-0015 | Agarose bead control |
| Bromophenol Blue | Fisher | BP112-25 | Component of SDS-PAGE loading buffer |
| 1.5 mL Centrifuge Tubes | FroggaBio | 1210-00S | Used to centrifuge small volumes |
| 15 mL Conical Centrifuge Tubes | Fisher | 1495970C | Used in culturing primary cells |
| Defined trypsin inhibitor | Fisher | R007100 | DTI |
| Dithiothreital | Fisher | BP172-25 | DTT |
| Epithelial cell medium (complete kit) | ScienCell | 4101 | Includes serum and growth factor supplements) |
| Glycerol | Fisher | BP229-1 | Component of SDS-PAGE loading buffer |
| 100 mM Guanosine 5'-triphosphate, 1 mL | Jena Bioscience | 272076-0251M | GTP |
| HCT116 colorectal carcinoma | ATCC | CCL-247 | Human cancer cell line |
| Human renal proximal tubular epithelial cells | ATCC | PCS-400-010 | HRPTEC |
| Hyclone DMEM/High Glucose | GE Life Sciences | SH30022.01 | Standard media for human cell culture |
| Hyclone Penicillin-Streptomycin solution | GE Life Sciences | SV30010 | Antibiotic component of DMEM |
| H35 HypOxystation | Hypoxygen | N/A | Hypoxia workstation |
| Igepal CA-630 | MP Biomedicals | 2198596 | Detergent component of lysis buffer |
| Monopotassium phosphate | Fisher | P288-500 | KH2PO4 |
| Potassium chloride | Fisher | P217-500 | KCl |
| Magnesium chloride | Fisher | M33-500 | MgCl2 |
| Sodium chloride | Fisher | BP358-10 | NaCl |
| Sodium fluoride | Fisher | 5299-100 | NaF (phosphatase inhibitor component of lysis buffer) |
| Disodium phosphate | Fisher | 5369-500 | Na2HPO4 |
| Premium Grade Fetal Bovine Serum | Seradigm | 1500-500 | FBS |
| Protease Inhibitor Cocktail (100 x) | Cell Signalling | 58715 | Component of lysis buffer |
| Sodium Dodecyl Sulfate | Fisher | BP166-100 | SDS |
| Sodium Orthovanadate | Sigma | 56508 | Na3VO4 |
| Tris Base | Fisher | BP152-5 | Component of buffers |
| 0.05% Trypsin-EDTA (1x) | Life Technologies | 2500-067 | Trypsin used to detach adherent cells |
Referencias
- Holcik, M., Sonenberg, N. Translational control in stress and apoptosis. Nat Rev Mol Cell Biol. 6 (4), 318-327 (2005).
- Sonenberg, N., Hinnebusch, A. G. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 136 (4), 731-745 (2009).
- Gingras, A. C., Raught, B., Sonenberg, N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu Rev Biochem. 68, 913-963 (1999).
- Weingarten-Gabbay, S., et al. Comparative genetics. Systematic discovery of cap-independent translation sequences in human and viral genomes. Science. 351 (6270), (2016).
- Andreev, D. E., et al. Oxygen and glucose deprivation induces widespread alterations in mRNA translation within 20 minutes. Genome Biol. 16, 90 (2015).
- Ho, J. J., et al. Systemic Reprogramming of Translation Efficiencies on Oxygen Stimulus. Cell Rep. 14 (6), 1293-1300 (2016).
- Shatsky, I. N., Dmitriev, S. E., Andreev, D. E., Terenin, I. M. Transcriptome-wide studies uncover the diversity of modes of mRNA recruitment to eukaryotic ribosomes. Crit Rev Biochem Mol Biol. 49 (2), 164-177 (2014).
- Ho, J. J., Lee, S. A Cap for Every Occasion: Alternative eIF4F Complexes. Trends Biochem Sci. , (2016).
- Lin, T. A., et al. PHAS-I as a link between mitogen-activated protein kinase and translation initiation. Science. 266 (5185), 653-656 (1994).
- Richter, J. D., Sonenberg, N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 433 (7025), 477-480 (2005).
- Tee, A. R., Tee, J. A., Blenis, J. Characterizing the interaction of the mammalian eIF4E-related protein 4EHP with 4E-BP1. FEBS Lett. 564 (1-2), 58-62 (2004).
- Rom, E., et al. Cloning and characterization of 4EHP, a novel mammalian eIF4E-related cap-binding protein. J Biol Chem. 273 (21), 13104-13109 (1998).
- Uniacke, J., et al. An oxygen-regulated switch in the protein synthesis machinery. Nature. 486 (7401), 126-129 (2012).
- Morita, M., et al. A novel 4EHP-GIGYF2 translational repressor complex is essential for mammalian development. Mol Cell Biol. 32 (17), 3585-3593 (2012).
- Webb, N. R., Chari, R. V., DePillis, G., Kozarich, J. W., Rhoads, R. E. Purification of the messenger RNA cap-binding protein using a new affinity medium. Bioquímica. 23 (2), 177-181 (1984).
- Kiriakidou, M., et al. An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell. 129 (6), 1141-1151 (2007).
- Mazza, C., Segref, A., Mattaj, I. W., Cusack, S. Large-scale induced fit recognition of an m(7)GpppG cap analogue by the human nuclear cap-binding complex. EMBO J. 21 (20), 5548-5557 (2002).
- Nojima, T., Hirose, T., Kimura, H., Hagiwara, M. The interaction between cap-binding complex and RNA export factor is required for intronless mRNA export. J Biol Chem. 282 (21), 15645-15651 (2007).
- Pabis, M., Neufeld, N., Shav-Tal, Y., Neugebauer, K. M. Binding properties and dynamic localization of an alternative isoform of the cap-binding complex subunit CBP20. Nucleus. 1 (5), 412-421 (2010).
- Ruud, K. A., Kuhlow, C., Goss, D. J., Browning, K. S. Identification and characterization of a novel cap-binding protein from Arabidopsis thaliana. J Biol Chem. 273 (17), 10325-10330 (1998).
- Ptushkina, M., et al. A second eIF4E protein in Schizosaccharomyces pombe has distinct eIF4G-binding properties. Nucleic Acids Res. 29 (22), 4561-4569 (2001).
- Cho, P. F., et al. A new paradigm for translational control: inhibition via 5′-3′ mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell. 121 (3), 411-423 (2005).
- Dinkova, T. D., Keiper, B. D., Korneeva, N. L., Aamodt, E. J., Rhoads, R. E. Translation of a small subset of Caenorhabditis elegans mRNAs is dependent on a specific eukaryotic translation initiation factor 4E isoform. Mol Cell Biol. 25 (1), 100-113 (2005).
- Timpano, S., Uniacke, J. Human Cells Cultured Under Physiological Oxygen Utilize Two Cap-binding Proteins to Recruit Distinct mRNAs for Translation. J Biol Chem. , (2016).
- Dings, J., Meixensberger, J., Jager, A., Roosen, K. Clinical experience with 118 brain tissue oxygen partial pressure catheter probes. Neurosurgery. 43 (5), 1082-1095 (1998).
- Le, Q. T., et al. An evaluation of tumor oxygenation and gene expression in patients with early stage non-small cell lung cancers. Clin Cancer Res. 12 (5), 1507-1514 (2006).
- Muller, M., et al. Effects of desflurane and isoflurane on intestinal tissue oxygen pressure during colorectal surgery. Anaesthesia. 57 (2), 110-115 (2002).
- Brooks, A. J., Eastwood, J., Beckingham, I. J., Girling, K. J. Liver tissue partial pressure of oxygen and carbon dioxide during partial hepatectomy. Br J Anaesth. 92 (5), 735-737 (2004).
- Muller, M., et al. Renocortical tissue oxygen pressure measurements in patients undergoing living donor kidney transplantation. Anesth Analg. 87 (2), 474-476 (1998).
- Richardson, R. S., et al. Human skeletal muscle intracellular oxygenation: the impact of ambient oxygen availability. J Physiol. 571 (Pt 2), 415-424 (2006).
- Harrison, J. S., Rameshwar, P., Chang, V., Bandari, P. Oxygen saturation in the bone marrow of healthy volunteers. Blood. 99 (1), 394 (2002).
- Gleadle, J., Ratcliffe, P. . Hypoxia. , (2001).
- Gluckman, E., et al. Hematopoietic reconstitution in a patient with Fanconi’s anemia by means of umbilical-cord blood from an HLA-identical sibling. N Engl J Med. 321 (17), 1174-1178 (1989).
- Parrinello, S., et al. Oxygen sensitivity severely limits the replicative lifespan of murine fibroblasts. Nat Cell Biol. 5 (8), 741-747 (2003).
- Jewell, U. R., et al. Induction of HIF-1alpha in response to hypoxia is instantaneous. FASEB J. 15 (7), 1312-1314 (2001).
- Newby, D., Marks, L., Lyall, F. Dissolved oxygen concentration in culture medium: assumptions and pitfalls. Placenta. 26 (4), 353-357 (2005).
- Towbin, H., Staehelin, T., Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 76 (9), 4350-4354 (1979).
- Haghighat, A., Mader, S., Pause, A., Sonenberg, N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 14 (22), 5701-5709 (1995).
- Pyronnet, S., et al. Human eukaryotic translation initiation factor 4G (eIF4G) recruits mnk1 to phosphorylate eIF4E. EMBO J. 18 (1), 270-279 (1999).
- Okumura, F., Zou, W., Zhang, D. E. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 21 (3), 255-260 (2007).
- Kedersha, N., et al. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol Biol Cell. 13 (1), 195-210 (2002).
- Gu, M., et al. Insights into the structure, mechanism, and regulation of scavenger mRNA decapping activity. Mol Cell. 14 (1), 67-80 (2004).
- Szczepaniak, S. A., Zuberek, J., Darzynkiewicz, E., Kufel, J., Jemielity, J. Affinity resins containing enzymatically resistant mRNA cap analogs–a new tool for the analysis of cap-binding proteins. RNA. 18 (7), 1421-1432 (2012).
- Joshi, B., Cameron, A., Jagus, R. Characterization of mammalian eIF4E-family members. Eur J Biochem. 271 (11), 2189-2203 (2004).

