Immunofluorescence Analysis of Stress Granule Formation After Bacterial Challenge of Mammalian Cells
Summary
We describe a method for the qualitative and quantitative analysis of stress granule formation in mammalian cells after the cells are challenged with bacteria and a number of different stresses. This protocol can be applied to investigate the cellular stress granule response in a wide range of host-bacterial interactions.
Abstract
Fluorescent imaging of cellular components is an effective tool to investigate host-pathogen interactions. Pathogens can affect many different features of infected cells, including organelle ultrastructure, cytoskeletal network organization, as well as cellular processes such as Stress Granule (SG) formation. The characterization of how pathogens subvert host processes is an important and integral part of the field of pathogenesis. While variable phenotypes may be readily visible, the precise analysis of the qualitative and quantitative differences in the cellular structures induced by pathogen challenge is essential for defining statistically significant differences between experimental and control samples. SG formation is an evolutionarily conserved stress response that leads to antiviral responses and has long been investigated using viral infections1. SG formation also affects signaling cascades and may have other still unknown consequences2. The characterization of this stress response to pathogens other than viruses, such as bacterial pathogens, is currently an emerging area of research3. For now, quantitative and qualitative analysis of SG formation is not yet routinely used, even in the viral systems. Here we describe a simple method for inducing and characterizing SG formation in uninfected cells and in cells infected with a cytosolic bacterial pathogen, which affects the formation of SGs in response to various exogenous stresses. Analysis of SG formation and composition is achieved by using a number of different SG markers and the spot detector plug-in of ICY, an open source image analysis tool.
Introduction
Visualizing host-pathogen interactions on a cellular level is a powerful method for gaining insights into pathogenic strategies and for identifying key cellular pathways. Indeed, pathogens can be used as tools to pinpoint important cellular targets or structures, as pathogens have evolved to subvert central cellular processes as a strategy for their own survival or propagation. Visualization of cellular components can be achieved by recombinantly expressing fluorescently-tagged host proteins. While this allows for real-time analysis, the generation of cell lines with specifically-tagged host proteins is highly laborious and may result in undesirable side effects. More convenient is the detection of cellular factors using specific antibodies, because multiple host factors can be analyzed simultaneously and one is not limited to a particular cell type. A drawback is that only a static view can be captured as immunofluorescence analysis necessitates host cell fixation. However, an important advantage of immunofluorescence imaging is that it readily lends itself to both qualitative and quantitative analysis. This in turn can be used to obtain statistically significant differences to provide new insights into host-pathogen interactions.
Fluorescent image analysis programs are powerful analytical tools for performing 3D and 4D analysis. However, the high cost of software and its maintenance make methods based on free open source software more widely attractive. Careful image analysis using bio-analysis software is valuable as it substantiates visual analysis and, when assigning statistical significances, increases confidence in the correctness of a given phenotype. Previously, SGs have been analyzed using the free ImageJ software, which necessitates the manual identification of individual SGs4. Here we provide a protocol for the induction and analysis of cellular SG formation in the context of bacterial infections using the free open source bio-image analysis software ICY (http://icy.bioimageanalysis.org). The bio-image analysis software has a built-in spot detector program that is highly suitable for SG analysis. It allows the fine-tuning of the automated detection process in specified Regions Of Interest (ROIs). This overcomes the need for manual analysis of individual SGs and removes sampling bias.
Many environmental stresses induce the formation of SGs, which are phase dense cytosolic, non-membranous structures of 0.2 – 5 µm in diameter5,6. This cellular response is evolutionarily conserved in yeast, plants and mammals and occurs when global protein translation is inhibited. It involves aggregation of stalled translation initiation complexes into SGs, which are considered holding places for translationally-inactive mRNAs, allowing selective translation of a subset of cellular mRNAs. Upon removal of the stress, SGs dissolve and global rates of protein synthesis resume. SGs are composed of translation elongation initiation factors, proteins involved in RNA metabolism, RNA-binding proteins, as well as scaffolding proteins and factors involved in host cell signaling2, although the exact composition can vary depending on the stress applied. Environmental factors that induce SG formation include amino acid starvation, UV irradiation, heat shock, osmotic shock, endoplasmic reticulum stress, hypoxia and viral infection2,7,8. Much progress has been made in understanding how viruses induce and also subvert SG formation, while little is still known about how other pathogens, such as bacterial, fungal or protozoan pathogens, affect this cellular stress response1,7.
Shigella flexneri is a gram-negative facultative cytosolic pathogen of humans and the causative agent of severe diarrhea or shigellosis. Shigellosis is a major public health burden and leads to 28,000 deaths annually in children under 5 years of age9,10. S. flexneri infects the colonic epithelium and spreads cell-to-cell by hijacking the host's cytoskeletal components11,12. Infection of the epithelium supports the replication of S. flexneri within the cytosol but infected macrophages die through an inflammatory cell death process called pyroptosis. Infection leads to a massive recruitment of neutrophils and severe inflammation that is accompanied by heat, oxidative stress and tissue destruction. Thus, while infected cells are subject to internal stresses induced by infection, such as Golgi disruption, genotoxic stress and cytoskeletal rearrangements, infected cells are also subjected to environmental stresses due to the inflammatory process.
Characterization of the effect of S. flexneri infection on the ability of cells to respond to environmental stresses using a number of SG markers has demonstrated that infection leads to qualitative and quantitative differences in SG composition3. However, little is known about other bacterial pathogens. Here we describe a methodology for the infection of host cells with the cytosolic pathogen S. flexneri, the stressing of cells with different environmental stresses, the labeling of SG components, and the qualitative and quantitative analysis of SG formation and composition in the context of infected and non-infected cells. This method is widely applicable to other bacterial pathogens. In addition, the image analysis of the SG formation may be used for infections by viruses or other pathogens. It can be used to analyze SG formation upon infection or the effect of infection on SG formation in response to exogenous stresses.
Protocol
1. Preparation of Bacteria and Host Cells
- Transfer 12 or 14 mm glass cover slips in either 24- or 12-well plates, respectively, with sterile tweezers, counting one well per experimental condition. Sterilize these by UV treatment in a tissue culture hood for 15 min.
NOTE: Include control wells for bacterial challenge and for SG induction by exogenous stresses. - For a 24-well plate with 12 mm coverslips, seed 1 x 105 mammalian cells (e.g., HeLa cells) onto coverslips; press the coverslips down with a sterile pipette tip. Let cells adhere overnight at 37 °C in a 5% CO2 tissue culture incubator in culture medium appropriate for each cell type.
NOTE: Plate cells so that the confluence of cells is between 40 – 60% the following day.- For HeLa cells, incubate in Dulbecco's Modified Eagle Medium (DMEM) with Non-Essential Amino Acids (NEAA) and 10% decomplemented Fetal Calf Serum (FCS). Decomplement FCS by heating for 30 min in a 56 °C water bath, swirling the serum once after 15 min.
- To streak out bacteria, use a sterile pipette tip to remove a small amount from a bacterial glycerol stock (20% glycerol) stored in a -80 °C freezer and place it on a labeled plate. Use a sterile loop to spread the bacteria over about 10% of the plate. Use a new sterile loop and drag it through the smear to make 3 parallel lines, half a plate long. Streak through these lines covering the rest of the plate. Incubate plate O/N at 37 °C.
- Select a single bacterial colony (e.g., S. flexneri) from the freshly streaked-out plate. Avoid working with bacterial colonies older than 1 week.
- For S. flexneri strain M90T, inoculate a red colony from a freshly streaked out Tryptic Soy Agar-0.1% Congo red agar plate into 8 mL of Tryptic Soy broth in a 15 mL tube; tighten the lid and incubate shaking at 222 rpm O/N at 30 °C.
CAUTION: Do not use large white colonies as they have lost the virulence plasmid and are non-infective.
2. Bacterial Challenge of Host Cells
- Prepare bacteria to maximize infection potential.
- For S. flexneri, subculture bacteria by adding 150 µL O/N culture to 8 mL Tryptic Soy Agar and incubate shaking 222 rpm at 37 °C to late exponential phase (~2 h) to induce virulence gene expression and enhance infection.
- When the culture is at an optical density between 0.6 and 0.9 (measured at 600 nm), transfer 1 mL of culture into a 1.5 mL tube. Pellet bacteria for 2 min at 8,000 x g and resuspend in infection medium (DMEM with NEAA, lacking FCS) at OD600 = 1. Thus, if the OD600 is 0.85, resuspend in 850 µL.
NOTE: For S. flexneri, at OD600 = 1, there will be 8 x 108 bacteria per mL when cultured in 8 mL medium. A Multiplicity Of Infection (MOI) of 20 is appropriate. For other pathogens, the number of bacteria per mL at an OD600 and the appropriate MOI need to be determined empirically.
- When the culture is at an optical density between 0.6 and 0.9 (measured at 600 nm), transfer 1 mL of culture into a 1.5 mL tube. Pellet bacteria for 2 min at 8,000 x g and resuspend in infection medium (DMEM with NEAA, lacking FCS) at OD600 = 1. Thus, if the OD600 is 0.85, resuspend in 850 µL.
- To enhance bacterial-host interactions, coat bacteria with Poly-L-Lysine (PLL).
NOTE: PLL treatment of S. flexneri had no effect on SG dynamics. Other pathogens need to be assessed for their amenability to PLL treatment.- To coat bacteria with PLL, centrifuge 1 mL of bacterial culture for 2 min at 8,000 x g and then resuspend the pellet in 10 µg/mL PLL in Phosphate-Buffered Saline (PBS) at an OD600 = 1.
- Add the bacterial solution to a small dish, such as a well within a 12-well plate, and incubate at RT (~ 20 °C) for 10 min with gentle agitation on a plate shaker.
- Pellet bacteria for 2 min at 8,000 x g, remove the excess PLL. Resuspend the pellet in 1 mL PBS and repeat this washing step twice. After the final wash, resuspend in infection medium at OD600 = 1.
NOTE: Here, the infection medium for S. flexneri is host cell medium with 20 mM HEPES w/o FCS.
- For S. flexneri, subculture bacteria by adding 150 µL O/N culture to 8 mL Tryptic Soy Agar and incubate shaking 222 rpm at 37 °C to late exponential phase (~2 h) to induce virulence gene expression and enhance infection.
- Prepare cells for bacterial challenge.
- Remove medium from mammalian HeLa cells using a suction pump. Add 500 µL RT HeLa cell medium to wash the cells, swirl, remove medium using a suction pump, and replace with 500 µL RT appropriate infection medium (e.g., host cell medium with 20 mM HEPES w/o FCS).
- NOTE: For all washing steps, work quickly and process only up to 4 coverslips at a time to avoid drying out of the cells.
- Challenge prepared HeLa cells with bacteria to infect 20 – 50% of the cells.
NOTE: The appropriate time and MOI need to be determined for each bacterial species. Generally, a good start is 30 min at 37 °C with a MOI of 10.- For S. flexneri, challenge HeLa cells cells using one of the two methods (step 2.3.1.1 or 2.3.1.2).
- Spin non-PLL-treated bacteria (20 µL of OD600 = 1/well of 24-well plate in 500 µL medium) onto cells for 10 min at 13000 x g.
- Add PLL-coated bacteria (15 µL of OD600 = 1/well of 24-well plate in 500 µL medium) for 15 min at RT to allow bacteria to settle onto cells.
- Allow the infection to occur by placing cells with settled bacteria into a 37 °C tissue culture incubator for 30 min.
NOTE: For short time points, synchronize S. flexneri infection by placing plates in a 37 °C water bath for 15 min instead of in the tissue culture incubator.
- For S. flexneri, challenge HeLa cells cells using one of the two methods (step 2.3.1.1 or 2.3.1.2).
- Stop infection by washing cells.
- To wash cells and remove excess bacteria, remove the medium using a suction pump, add 1 mL fresh prewarmed (37 °C) HeLa culture medium (DMEM, 10% FCS, NEAA). Swirl the medium, remove and replace with fresh medium. Repeat twice and leave fresh medium on the cells.
- For S. flexneri, or other intracellular bacteria, after infection of HeLa cells and the washes, replace medium with standard tissue culture medium containing 50 µg/mL gentamicin to kill extracellular bacteria and prevent further cell invasion.
NOTE: Do not add antibiotics in the medium for extracellular bacteria.
- For S. flexneri, or other intracellular bacteria, after infection of HeLa cells and the washes, replace medium with standard tissue culture medium containing 50 µg/mL gentamicin to kill extracellular bacteria and prevent further cell invasion.
- To wash cells and remove excess bacteria, remove the medium using a suction pump, add 1 mL fresh prewarmed (37 °C) HeLa culture medium (DMEM, 10% FCS, NEAA). Swirl the medium, remove and replace with fresh medium. Repeat twice and leave fresh medium on the cells.
- Incubate bacteria with cells for the desired amount of time in a tissue culture incubator.
NOTE: For S. flexneri, infection may be as long as 6 h depending on the cell type. For HeLa cells, let infection proceed for 1.5 – 2 h before adding exogenous stresses to induce SG formation. For Caco-2 infections, in which Shigella spreads cell-to-cell, infect for 30 min to 6 h before adding exogenous stress. Cells may be fixed at this point without adding exogenous stresses to assess the formation of SGs as a result of the bacterial infection (in that case, proceed to section 4).
3. Inducing Stress Granule Formation by the Addition of Exogenous Stressors
- Remove the medium from infected and non-infected HeLa cells using a suction pump, replace medium with 500 µL of appropriate medium with or without the stressor and incubate.
NOTE: Stress-inducing conditions include the following (see below). For additional treatments see Kedersha et al.13.- For heat shock treatment, use regular medium containing 20 mM HEPES and place the plate into 44 °C water bath for 30 min.
- For clotrimazole (induces mitochondrial stress) treatment, use regular medium without FCS with 20 µM clotrimazole and incubate in a tissue culture incubator for 1 h.
NOTE: Store single-use aliquots of 20 mM clotrimazole stock in dimethyl sulfoxide (DMSO) at -20 °C for up to 4 months. Use DMSO vehicle for control cells. - For arsenite (induces oxidative stress) treatment, use regular medium with 0.5 µM arsenite and incubate in tissue culture incubator for 1 h.
NOTE: Store single-use aliquots of 0.5 mM arsenite stock at -20 °C for up to 6 months.
CAUTION: Clotrimazole and arsenite are harmful and dangerous to the environment and must be disposed of appropriately. - For Des-Methyl Des-Amino (DMDA)-pateamine treatment (inhibits eukaryotic translation initiation), use regular medium with 50 – 200 nM DMDA-pateamine and incubate in tissue culture incubator for 1 h.
NOTE: Store DMDA-pateamine at -80 °C. Store reagents as small aliquots to avoid freeze thawing.
4. Fixation and Immunofluorescence Analysis of Stress Granule Formation
NOTE: Process the control and experimental coverslips at the same time to avoid staining differences that may impact image analysis in subsequent steps. The control samples include no infection with and without SG inducing treatment, and infected samples with and without SG inducing treatment.
- Remove medium completely using a suction pump and fix samples by adding 0.5 mL RT 4% paraformaldehyde (PFA) in PBS. Incubate for 30 min at RT.
CAUTION: PFA is harmful to the handler and the environment. Take proper measure to protect the handler and dispose with chemical waste.
NOTE: Alternatively, fix for 15 min and then replace with -20 °C prechilled methanol for 10 min to retain more cytoplasmic localization of SG markers13. - Remove the PFA with a pipette and dispose the liquid in an appropriate waste container. Wash the coverslip with 1 mL Tris-Buffered Saline (TBS: 25 mM Tris, pH 7.3; 15 mM NaCl). Incubate the coverslip for 2 min with gentle shaking on a plate shaker during washing.
- Remove TBS completely using a suction pump and add 500 µL 0.3% Triton-X100 in TBS for 10 min to permeabilize the cells.
- Remove permeabilizing solution with a suction pump. Replace with 1 mL TBS, swirl the plate gently, remove TBS by suction, and add 1 mL blocking solution (5% FCS in TBS) for 1 h at RT.
- Prepare the primary antibody solution (1:300 dilution) in 5% FCS in TBS. Calculate 50 µL per 12 mm coverslip and make enough for all coverslips in one batch.
NOTE: The common primary antibodies that are used target G3BP1, eIF3b, and TIA1. - Spot 50 µL of the primary antibody mix onto a plastic paraffin film (parafilm) firmly taped onto the bench or chamber.
NOTE: A list of canonical SG markers are provided in the Table of Materials, but composition of SGs may depend on the stress applied. Other SG markers are listed in Kedersha et al.13 Expected results for a number of SG markers for S. flexneri infection are listed in Table 1. - Pick up the coverslip with tweezers, dab the edge of the coverslip onto tissue, briefly releasing the tweezers to remove excess liquid, and gently place face down onto the drop. Incubate coverslips for 1.5 h at RT or overnight at 4 °C in a moist chamber.
NOTE: To prepare a moist chamber, place prewetted tissues along the edges of a firmly closed chamber lined with plastic parafilm. - Transfer each coverslip with tweezers into a well of a new 24-well plate filled with 1 mL TBS; incubate with gentle shaking on a plate shaker for 5 min. Suction off TBS in each well using a suction pump, replace with 1 mL TBS and place back on plate shaker for 5 min. Repeat wash one more time.
NOTE: Reuse the parafilm by removing the excess liquid with a tissue and washing the surface with water. Ensure the paraffin film is completely dry before using it for the subsequent staining step. - Prepare the secondary antibody solution in TBS (dilution 1:500 – 1:2,000). To stain the nucleus and bacteria, add 2 µg/mL DAPI to the mix. To stain actin, add fluorescent phalloidin. Pipette 50 µL secondary antibody mix onto the plastic paraffin film.
NOTE: Use of highly purified cross-absorbed donkey secondary antibodies are recommended for co-localization studies of different SG markers when G3BP1 (goat antibody) is used. Choose fluorescently labeled secondary antibodies with non-overlapping spectra. eIF3b clearly stains the cytoplasm of cells and is therefore a good marker to delineate cells. The actin stain further helps to delineate cells at cell-to-cell contact sites and areas of bacterial entry. - Pick up the coverslip with tweezers, dab the edge of coverslip onto tissue, releasing briefly the tweezers, to remove excess liquid. Gently place coverslip face down onto the drop and incubate the coverslips in the dark for 1 h at RT.
- Use tweezers to place the coverslip back into the 24-well plates filled with fresh 1 mL PBS. Ensure the coverslip is fully covered by the PBS. Wash the cells 3x with gentle shaking for 5 min each.
NOTE: Keep the plate during the washings under a cover to protect from light as the fluorophores of the secondary antibody are light sensitive. - Briefly dip the coverslip into deionized water to remove the salt, dab dry from the edge onto a tissue and mount the coverslip onto 5 µL of fluorescence mounting medium on a glass slide. Seal the coverslip prior to imaging using nail polish. Keep slides at 4 °C for short-term (week) or at -20 °C for long-term (month) storage.
NOTE: Avoid air bubbles when sealing as this will harm the image quality.
5. Fluorescence Imaging
NOTE: Refer to the user manual of the microscope for optimization of the set up.
- Acquire images using a confocal microscope using either a 40 or 63X oil immersion lens. Select the desired fluorescent channels for image acquisition. When using multiple fluorescent channels, select sequential acquisition mode.
NOTE: Start with the experimental samples where SGs have been induced the strongest to avoid overexposure on subsequent samples. Make sure to not have overexposed SGs throughout the entire depth of the cell.- Set bit size of 12 or higher within the microscope settings to ensure accuracy of the image analysis.
- Set the image stacks to cover the whole depth of the cell. Check in the different channels and for different SG markers to ensure that the whole range is included. Set the beginning of the stack at the base of the cells and the top of the stacks at the height at the top of the cells where no more SG markers are observed.
- Acquire the stack. Keep the stack height the same for all images.
NOTE: Do not change acquisition settings between control and experimental samples that will used for subsequent comparison.
6. Image Analysis
NOTE: Here, SG analysis on collapsed stacks using freeware ICY is described. Image analysis of 3D reconstructions may also be done using other specialized software. SG detection may be performed via a fully automated workflow established for this protocol (available at http://icy.bioimageanalysis.org/protocol/Stress_granule_detection_in_fluorescence_imaging). To use this protocol, the nuclei need to be stained with a DNA stain for the software to find the center of each cell, and the cell edges need to be marked by either a cytoplasmic marker (such as eIF3b) or actin stain for the software to identify the cell boundaries. The automatic workflow can be used to validate manually-derived results or directly for analysis when cell boundaries are detected with high confidence, which will mostly depend on the density of cells and the marker used to detect the cell boundaries.
- Using ImageJ or Fiji, collapse the image stacks (Image>Stacks>Z projection), and save as .tiff files.
NOTE: Create a new folder for each image as the image analysis software will link its results to the site of the original image. - Load a control and experimental .tiff image to the image analysis platform (File>Open).Go to the Sequence window. Scroll down in the "Lookup Table" to the channel parameters.
- Deactivate all channels by clicking on the check box of all channel tabs. Click on channel 0 and then select the preferred color by clicking on the color bar above. Increase the intensity of the channel as needed to see all structures by clicking and moving the line in the intensity field. Repeat this for all channels.
NOTE: Deactivate channels that are not required for a given task to minimize bias in sample analysis. - Scroll up and within the Canvas tab, zoom in to about 10 cells per field by adjusting the zoom tab.
- Use the polygon function tools (in the top bar) to delineate cell boundaries of cells in the image. Use the actin stain or a cytosolic marker for cell boundary delineation (e.g. eIF3b).
- Click the polygon function within the "File & ROI" tools. Start delineation by clicking on the cell and then extend the line by making anchors through repeated clicking around the cell boundary. To finish a ROI, click again on the polygon function within the "File & ROI" tools.
NOTE: Identify the sub-populations of the delineated cells that are infected. The sample consists of a mix of infected and non-infected cells. To identify bacteria, either use the DAPI stain or fluorescent bacteria (such as GFP-expressing bacteria). Increase the intensity to ensure that all bacteria are seen. - To name a ROI, go to the ROI window on the right and click on the cell of interest in the image. This will highlight the corresponding ROI in the ROI tab. Double click on the ROI name and modify the name (e.g. infected cell #1).
NOTE: If one image is to be used to analyze both infected and non-infected cells, it is easier to save the image in two separate folders and delineate the infected and non-infected cells separately, generating two different spreadsheets, which will facilitate analysis later.
- Click the polygon function within the "File & ROI" tools. Start delineation by clicking on the cell and then extend the line by making anchors through repeated clicking around the cell boundary. To finish a ROI, click again on the polygon function within the "File & ROI" tools.
- Open the spot detector application within the "Detection & Tracking" tab (top bar). Within the Input tab, the current image will be selected. Do not change anything. Within the Pre-Processing tab, select the channel to be analyzed.
NOTE: Only one channel can be analyzed at any given time. A batch analysis can be selected here if one uses the fully automated analysis sequence whose parameters have been checked to give satisfying results.- Within the Detector tab, chose "Detect bright spots over dark background". Then select the appropriate scale and sensitivity. Do this by testing different settings and cross-checking spot detection in both the control and the experimental samples.
NOTE: For an image with a 2,048 x 2,048 resolution and a pixel size of 0.12 µm2, a level 2 threshold with a sensitivity of 25 to 100 is generally appropriate but each SG marker has to be tested empirically. Set up spot detection so that in the non-treated control sample, spots are not detected, while in the experimental sample with SGs, all the small and large SGs are counted. - Within the ROI tab, select the suggested ROI From Sequence. Within the Filtering tab, select No Filtering. In the Output tab, choose the appropriate output.
NOTE: Choosing Enable Specific file is helpful for saving different detection conditions or different channels. Selecting to export the binary image and the original image with ROIs and detection is helpful for quality control analysis. - Select "Remove previous spots rendered as ROIs". Choose the folder to save the file by clicking on the "no file selected" button. Within the Display tab, select the desired options.Click "Start detection".
NOTE: A folder with the name and the location chosen will be created that contains the exported imaging options. These images will be overwritten for each new condition tested. To keep the images, either rename the folder so that a new folder will be created or transfer the images from the save folder to a new folder. For quality control of the images, it is helpful to select display detection mark, number of detection of ROI and ROI number and name.
- Within the Detector tab, chose "Detect bright spots over dark background". Then select the appropriate scale and sensitivity. Do this by testing different settings and cross-checking spot detection in both the control and the experimental samples.
- Consolidate and save the data for the different parameters using the generated spreadsheet for each image or condition tested.
NOTE: The parameters to analyze include the number of SGs per ROI, the SG surface areas, and the SG maximum, average and minimum intensities. - Transfer the data and analyze using an analysis program capable of analyzing, graphing and determining the statistical significance.
Representative Results
To explain and demonstrate the protocol described in this manuscript, we characterized the image of clotrimazole-induced SGs in HeLa cells infected or not with the cytosolic pathogen S. flexneri. An outline of the procedure is presented in Figure 1, and includes virulent and avirulent S. flexneri streaked on Congo Red plates, bacteria preparation, infection, addition of environmental stress, sample fixation and staining, sample imaging and quantitation, as well as image analysis. A number of different stresses may be used to induce SG formation and a variety of SG markers are available for interrogation. eIF3b is a canonical SG marker that also clearly stains the cytoplasmic compartment of the cell and can be used to identify cell edges. G3BP1 is a widely-used SG marker that aggregates into SGs without any background staining (Figure 2A, B & D). Cells are best imaged with a confocal microscope, and the stacks covering the whole cell depth are loaded onto the imaging analysis freeware ICY (Figure 2A – C). To better identify infected cells and to put cells into either the infected or non-infected group for later analysis, it is useful to increase the intensity of the nucleic acid stain (Figure 2D). Within the imaging analysis software, the spot detector is then used to identify SGs within each of the designated ROIs (Figure 2E) and a binary image is generated that displays all the identified SGs (Figure 2F).
SG analysis needs to be tailored to each SG marker analyzed and the appropriate setting for analysis must be carefully chosen. Focusing on the SG marker G3BP1, changing the size requirement of the spot detected or changing the sensitivity of the detection parameters will lead to varying results, as demonstrated in Figure 3A – C. The scale selection (1 – 3, although scales can be added) is based on pixel size and thus at higher scales, smaller SGs will not be counted and the numbers of SGs will decrease (Figure 3C). Sensitivity is measured from 1 – 100, with 100 being the most sensitive. By increasing the sensitivity, the number of SGs detected increases (Figure 3C). Thus, the correct settings must be found to minimize false positives and false negatives. This is best done by carefully analyzing the visuals obtained for each setting. For G3BP1, a scale of 2 with a sensitivity of 100 (2 – 100, highlighted in red) gave the best result. A scale of 2 with a lower sensitivity (50 or 25) left small SGs uncounted (see orange arrows). Similarly, a scale of 3 left small SG unaccounted while a scale of 1 oversampled. Importantly, additional scales can be added to focus only on large SGs, if this is warranted. For eIF3b, a scale of 2 with a sensitivity of 55 (2 – 55) gave the best result for eIF3b (highlighted in red) although red arrows highlight either under or oversampling in a scale 2 – 50 & 2 – 55, respectively.
Once the parameters for SG analysis are properly set, the data can be analyzed in many ways (Figure 4). The spot detector gives the number of SGs detected within each ROI such that the uninfected and infected cells can be compared (Figure 4A). Similarly, the size, in this case number of pixels, is given from which the surface area can be calculated (Figure 4B). Frequency distribution plots are also useful to highlight shifts in the size of SGs found in different cell populations. Here, a complete absence of large SGs in S. flexneri infected cells, as well as a definite shift in the distribution becomes clearer (Figure 4C). In addition, the intensity (shown here is minimum and maximum intensity) provides information on the quality of the SGs analyzed. For S. flexneri infected cells, SGs are significantly less intense. These analyses provide statistically relevant information on both the qualitative and quantitative nature of SGs formed in response to exogenous stress when cells are infected with or without S. flexneri.
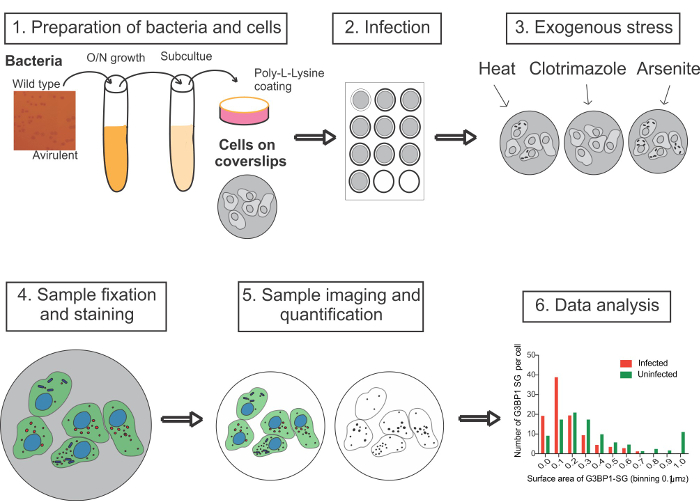
Figure 1: Outline of the Experimental Procedure to Infect Cells with S. Flexneri and to Analyze the Effect of Infection SG Formation. A Congo-red colony, and a nonvirulent Congo-red-negative colony, are picked and grown O/N in tryptic soy broth. The overnight culture is subcultured to the late exponential phase before bacteria are coated with PLL to promote adherence. Host cells grown on glass coverslips in 12-well plates are infected with bacteria for 30 min. Control cells are not infected. Cells are washed and treated with stress inducers to induce SG formation either by replacing the medium with stress-containing medium or subjecting the cells to stress conditions, such as heat. Cells are then fixed, permeabilized, stained with immunofluorescent SG-specific markers, and imaged using confocal microscopy. Z-projections are made using ImageJ and analyzed using the spot detector of the image analysis software. The data is then analyzed. Please click here to view a larger version of this figure.
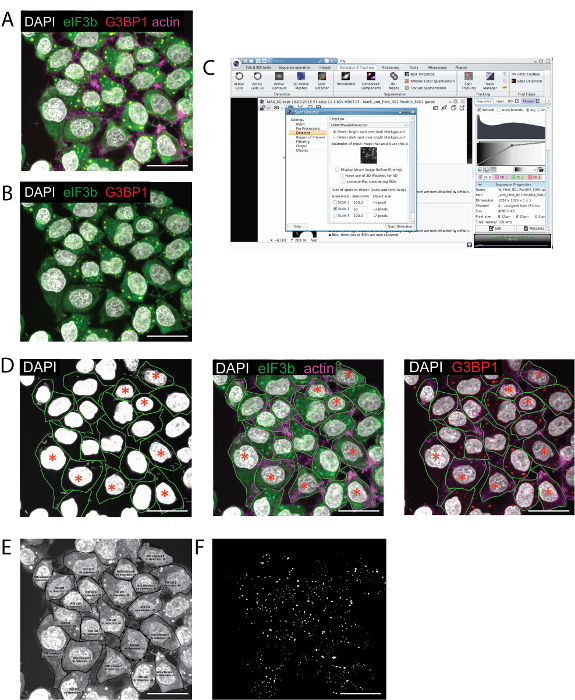
Figure 2: Spot Detector Analysis of HeLa Cells Infected with S. Flexneri for 1.5 h before the Addition of Clotrimazole for 1 h. A. Immunofluorescent image of a z-projection showing cells stained with the SG markers eIF3b and G3BP1, DAPI and actin. B. Same image as in (A) but without the actin stain to highlight the use of eIF3b to delineate cell boundaries. C. Screen shot of the image analysis software with the spot detector module. D. Gating of the infected and non-infected cells as seen using different channels. To see all bacteria, the intensity is increased. Cells with more than 1 bacterium present in the cell are labeled with a red star. E. Output of the spot detector analysis of individual ROIs, indicating the ROI name, the number of spots and their location. F. Image of spots detected in the ROIs. Scale bar = 30 µm. Please click here to view a larger version of this figure.
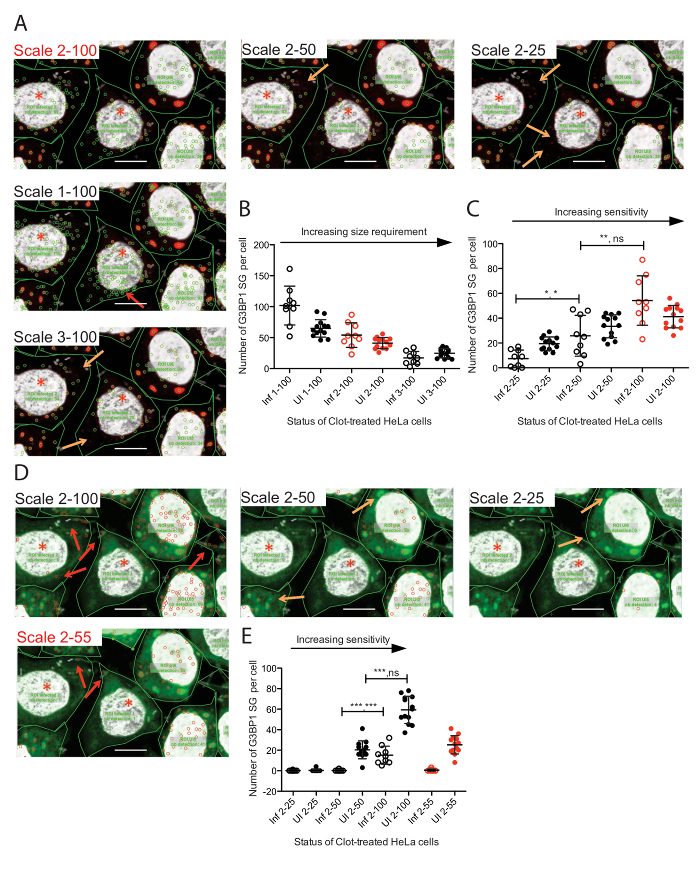
Figure 3: Comparisons of Different Scale and Sensitivity Settings of Spot Detector for the Detection of SGs through Different SG Markers. A. Zoom-in to HeLa cells infected or not with S. flexneri and stained with DAPI and the SG marker G3BP1, and analyzed at different scales (pixel size cut off, 1 – 3) and sensitivity (1 – 100). Graphical representation of SG numbers in uninfected and infected cells analyzed at different scales (B) and sensitivities (C). Visuals (D) and graphical representation (E) of SG number detection of the SG marker eIF3b for the same image when changing the sensitivity limit without changing the spot size cut-off. Red writing and red symbols indicate the best parameters. Red arrows point towards false positives and orange arrows to a lack of SG detection. Values include mean with standard deviation. Best results are highlighted in red. Selected statistical analysis included for clarity using the Wilcoxon rank sum test p-values on the left and the variance F-test on the right. * = <0.05, ** = <0.01, *** = <0.001, ns = nonsignificant. Scale bar = 10 µm. Please click here to view a larger version of this figure.
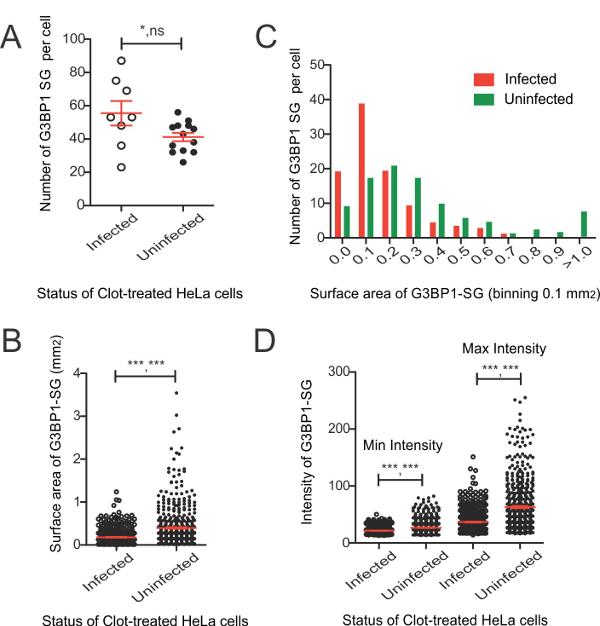
Figure 4: Analysis of Spot Detector Generated SG Data Obtained from Clotrimazole-treated HeLa Cells Infected with S. Flexneri. A. Number of G3BP1 SGs per cell in infected and uninfected HeLa cells. B. Surface area of G3BP1-SGs in infected and uninfected cells, and their percent distribution frequency (C). D. Minimum and maximum intensity values (arbitrary) of G3BP1-SGs. Values include mean with standard error. Selected statistical analysis included for clarity using the Wilcoxon rank sum test p-values on the left and the variance F-test on the right. * = <0.05, ** = <0.01, *** = <0.001, ns = nonsignificant. Please click here to view a larger version of this figure.
Discussion
The protocol outlined here describes the induction, localization, and analysis of SGs in non-infected cells and cells infected with the cytosolic pathogen S. flexneri in the presence or absence of exogenous stress. Using free imaging software, the protocols allows for the precise qualitative and quantitative analysis of SG formation to identify and statistically address differences in given phenotypes.
There are several critical steps within the protocol for the infection, SG-induction and imaging parts. For the infection, it is important that the bacterial interaction with host cells, such as invasion as outlined protocol here, is synchronized as much as possible. This is particularly important when investigating the effects of bacterial challenge early during the infection process. We previously showed that some phenotypes, such as eIF3b localization after S. flexneri infection, are perturbed early during wild-type S. flexneri challenge while other phenotypes, such as aggregation of G3BP1-containing SGs, are more affected at later time points3. Thus, to precisely describe the temporal effect of bacterial challenge, synchronization of infection is advisable. Notably, to analyze SG formation, cells should be in exponential phase of growth, and thus cell density at the time of stress addition is also an important parameter. It also limits SG analysis to non-confluent cell infection models. Another critical aspect is the co-processing of control and experimental samples during both the processing of the samples for imaging and during image acquisition. For immunofluorescent processing, all samples should be stained with the same antibody working dilutions for the same amount of time and under the same conditions (i.e. RT Vs. 4 °C), while also taking care to treat each coverslip similarly during the washing stages and mounting of the coverslips. This will ensure comparable immunofluorescent stainings that can be analyzed both qualitatively and quantitatively. Differences in mounting medium can have significant effects on bleaching of the samples during image acquisition and therefore should be kept constant. Similarly, image acquisition for all samples to be compared in analysis should be performed within one sitting and with the same settings to minimize differences arising from laser strength or sample set-up.
When using exogenous stresses, it is important to properly store the reagent and to frequently make new working stocks. Extended periods (>4 months for clotrimazole, for example) lead to decreased potency of the drug and will affect SG formation. Reagents should be added to culture medium immediately before addition to the cells; if a reduction in SG formation in control cells is observed or the aggregation of SGs appears aberrant, reagents should be tested for their activity and new stocks should be made.
One limitation is that SG identification using spot detector becomes less reliable when too much background fluorescence is present. While this is not a problem for many SG markers, some, such as eIF3b, which is a clear cytosolic marker under both SG-inducing and non-inducing conditions, are more difficult to analyze as demonstrated in Figure 3. In addition, scales and sensitivities need to be empirically determined for each SG marker. Another limitation is that the analysis through the image analysis software is best with 2D images, and therefore works well on optical slices or collapsed stacks, which however results in a loss of 3D information. The analysis on z-projections therefore tends to overstate the size of SGs and understate the number of SGs. 3D analysis of SGs may be performed with Imaris to give an even more precise spatial characterization, if deemed necessary for a particular phenotype.
SG characterization can be performed quickly and in a standardized manner using the automated spot detector of the image analysis software. In contrast, manual methods to identify and delineate SGs as previously performed4, leave the analysis open to more bias and is considerably more time consuming. SG analysis using spot detector can also be tailored to include or exclude small SG aggregates by selecting different sensitivity and size thresholds, thereby allowing flexibility in evaluating different aspects of SG formation. SG analysis can also be performed in a completely automated manner using an established automated workflow3. Within this workflow, the center of the cell is identified through a DAPI stain and the program then lets a malleable sphere grow outwards to delineate the cell boundaries based on either a cytoplasmic stain (such as eIF3b) or actin. At the same time, using a semi-automated analysis by manually delineating individual cells for analysis, can be advantageous when cells grow in clusters and cell boundaries are difficult for the automated program to delineate clearly. In these circumstances, defining cell boundaries manually can eliminate false positive or negative values.
The protocol may be adapted to other SG-inducing conditions and to other pathogens, including viruses, bacteria, yeast and protozoans. Of particular interest may be other cytosolic pathogens such as Rickettsia spp., Francisella tularensis, Burkholdieria pseudomallei and Listeria spp.14 For each infectious agent, and possibly also different cell lines, the infection protocol will have to be adapted and the sample times of exogenous stresses empirically determined. In addition, SG characterization using image analysis software may also be extended to live-cell imaging in the future. Real-time SG analysis would necessitate the expression of one or more fluorescently-tagged SG markers in the host cells but would thereby allow questions regarding the temporal and special dynamics of SG formation to be addressed under different experimental conditions. Fluorescently-labeled bacteria would then be needed to complement the real-time SG analysis in infected and uninfected cells.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
PS is a recipient of the Bill and Melinda Gates Grand Challenge Grant OPP1141322. PV was supported by a Swiss National Science Foundation Early Postdoc Mobility fellowship and a Roux-Cantarini postdoctoral fellowship. PJS is supported by an HHMI grant and ERC-2013-ADG 339579-Decrypt.
Materials
| Primary Antibodies | |||
| eIF3b (N20), origin goat | Santa Cruz | sc-16377 | Robust and widely used SG marker. Cytosolic staining allows cell delineation. Dilution 1 in 300 |
| eIF3b (A20), origin goat | Santa Cruz | sc-16378 | Same target as eIF3b (N20) and in our hands was identical to eIF3b (N20). Dilution 1 in 300 |
| eIF3A (D51F4), origin rabbit (MC: monoclonal) | Cell Signaling | 3411 | Part of multiprotein eIF3 complex with eIF3b . Dilution 1 in 800 |
| eIF4AI, origin goat | Santa Cruz | sc-14211 | Recommended by (Ref # 13). Dilution 1 in 200 |
| eIF4B, origin rabbit | Abcam | ab186856 | Good stress granule marker in our hands. Dilution 1 in 300 |
| eIF4B, origin rabbit | Cell Signaling | 3592 | Recommended by Ref # 13. Dilution 1 in 100 |
| eIF4G, origin rabbit | Santa Cruz | sc-11373 | Widely used SG marker. (Ref # 13): may not work well in mouse cell lines. Dilution 1 in 300 |
| G3BP1, origin rabbit (MC: monoclonal) | BD Biosciences | 611126 | Widely used SG marker. Dilution 1 in 300 |
| Tia-1, origin goat | Santa Cruz | sc-1751 | Widely used SG marker. Can also be found in P bodies when SG are present (Ref # 13). Dilution 1 in 300 |
| Alexa-conjugated Secondary Antibodies | |||
| A488 anti-goat , origin donkey | Thermo Fisher | A-11055 | Cross absorbed. Dilution 1 in 500 |
| A568 anti-goat, origin donkey | Thermo Fisher | A-11057 | Cross absorbed. Dilution 1 in 500 |
| A488 anti-mouse, origin donkey | Thermo Fisher | A-21202 | Dilution 1 in 500 |
| A568 anti-mouse, origin donkey | Thermo Fisher | A10037 | Dilution 1 in 500 |
| A647 anti-mouse, origin donkey | Thermo Fisher | A31571 | Dilution 1 in 500 |
| A488 anti-rabbit, origin donkey | Thermo Fisher | A-21206 | Dilution 1 in 500 |
| A568 anti-rabbit, origin donkey | Thermo Fisher | A10042 | Dilution 1 in 500 |
| Other Reagents | |||
| Shigella flexneri | Available from various laboratories by request | ||
| Tryptone Casein Soya (TCS) broth | BD Biosciences | 211825 | Standard growth medium for Shigella, application – bacterial growth |
| TCS agar | BD Biosciences | 236950 | Standard growth agar for Shigella, application – bacterial growth |
| Congo red | SERVA Electrophoresis GmbH | 27215.01 | Distrimination tool for Shigell that have lost the virulence plasmid, application – bacterial growth |
| Poly L lysine | Sigma-Aldrich | P1274 | Useful to coating bacteria to increase infection, application – infection |
| Gentamicin | Sigma-Aldrich | G1397 | Selective killing of extracellular but not cytosolic bacteria, application – infection |
| HEPES | Life Technologies | 15630-056 | PH buffer useful when cells are incubated at room-temperature, application – cell culture |
| DMEM | Life Technologies | 31885 | Standard culture medium for HeLa cells, application – cell culture |
| Fetal calf serum | Biowest | S1810-100 | 5% supplementation used for HeLa cell culture medium, application – cell culture |
| Non-essential amino acids | Life Technologies | 11140 | 1/100 dilution used for HeLa cell culture medium, application – cell culture |
| DMSO | Sigma-Aldrich | D2650 | Reagent diluent, application – cell culture |
| Sodium arsenite | Sigma-Aldrich | S7400 | Potent stress granule inducer (Note: highly toxic, special handling and disposal required), application – stress inducer |
| Clotrimazole | Sigma-Aldrich | C6019 | Potent stress granule inducer (Note:health hazard, special handling and disposal required), application – stress inducer |
| PFA | Electron Microscopy Scences | 15714 | 4% PFA is used for standard fixation of cells, application – fixation |
| Triton X-100 | Sigma-Aldrich | T8787 | Used at 0.03% for permeabilizationof host cells before immunofluorescent staining, application – permeabilization |
| A647-phalloidin | Thermo Fisher | A22287 | Dilution is at 1/40, best added during 2ary antibody staining, application – staining |
| DAPI | Sigma-Aldrich | D9542 | Nucleid acid stain used to visualize both the host nucleus and bacteria, application – staining |
| Parafilm | Sigma-Aldrich | BR701501 | Paraffin film useful for immunofluorescent staining of coverslips, application – staining |
| Prolong Gold | Thermo Fisher | 36930 | Robust mounting medium that works well for most fluorophores , application – mounting |
| Mowiol | Sigma-Aldrich | 81381 | Cheap and robust mounting medium that works well for most fluorophores, application – mounting |
| 24-well cell culture plate | Sigma-Aldrich | CLS3527 | Standard tissue culture plates, application – cell culture |
| 12-mm glass coverslips | NeuVitro | 1001/12 | Cell culture support for immunofluorescent applications, application – cell support |
| forceps | Sigma-Aldrich | 81381 | Cheap and obust mounting medium that works well for most fluorophores, application – mounting |
| Programs and Equipment | |||
| Prism | GraphPad Software | Data analysisand graphing program with robust statistical test options, application – data analysis | |
| Leica SP5 | Leica Microsystems | Confocal microsope, application – image acquisition | |
| Imaris | Bitplane | Professional image analysis program, application – data analysis | |
| Excel | Microsoft | Data analysis and graphing program, application – data analysis | |
Referencias
- Reineke, L. C., Lloyd, R. E. Diversion of stress granules and P-bodies during viral infection. Virology. 436 (2), 255-267 (2013).
- Kedersha, N., Ivanov, P., Anderson, P. Stress granules and cell signaling: more than just a passing phase. Trends Biochem Sci. 38 (10), 494-506 (2013).
- Vonaesch, P., Campbell-Valois, F. -. X., Dufour, A., Sansonetti, P. J., Schnupf, P. Shigella flexneri modulates stress granule composition and inhibits stress granule aggregation. Cell Microbiol. 18 (7), 982-997 (2016).
- Kolobova, E., Efimov, A., et al. Microtubule-dependent association of AKAP350A and CCAR1 with RNA stress granules. Exp Cell Res. 315 (3), 542-555 (2009).
- Anderson, P., Kedersha, N. Stress granules: the Tao of RNA triage. Trends in biochemical sciences. 33 (3), 141-150 (2008).
- Anderson, P., Kedersha, N. Stressful initiations. J Cell Sci. 115, 3227-3234 (2002).
- Mohr, I., Sonenberg, N. Host translation at the nexus of infection and immunity. Cell Host Microbe. 12 (4), 470-483 (2012).
- Hu, S., Claud, E. C., Musch, M. W., Chang, E. B. Stress granule formation mediates the inhibition of colonic Hsp70 translation by interferon- and tumor necrosis factor. Am J Physiol Gastointest Liver Physiol. 298 (4), 481-492 (2010).
- Barry, E. M., Pasetti, M. F., Sztein, M. B., Fasano, A., Kotloff, K. L., Levine, M. M. Progress and pitfalls in Shigella vaccine research. Nat Rev Gastroenterol Hepatol. 10 (4), 245-255 (2013).
- Lanata, C. F., Fischer-Walker, C. L., et al. Global Causes of Diarrheal Disease Mortality in Children. PloS One. 8 (9), 72788 (2013).
- Ashida, H., Ogawa, M., et al. Shigella deploy multiple countermeasures against host innate immune responses. Curr Opin Microbiol. 14 (1), 16-23 (2011).
- Ashida, H., Ogawa, M., Mimuro, H., Kobayashi, T., Sanada, T., Sasakawa, C. Shigella are versatile mucosal pathogens that circumvent the host innate immune system. Curr Opin Immunol. 23 (4), 448-455 (2011).
- Kedersha, N., Anderson, P. Mammalian stress granules and processing bodies. Methods Enzymol. 431, 61-81 (2007).
- Ray, K., Marteyn, B., Sansonetti, P. J., Tang, C. M. Life on the inside: the intracellular lifestyle of cytosolic bacteria. Nat Re. Micro. 7 (5), 333-340 (2009).

