In Vitro Culture of Epithelial Cells from Different Anatomical Regions of the Human Amniotic Membrane
Summary
This protocol describes the isolation of epithelial cells from different anatomical regions of the human amniotic membrane to determine their heterogeneity and functional properties for possible application in clinical and physiopathological models.
Abstract
Several protocols have been reported in the literature for the isolation and culture of human amniotic epithelial cells (HAEC). However, these assume that the amniotic epithelium is a homogeneous layer. The human amnion can be divided into three anatomical regions: reflected, placental, and umbilical. Each region has different physiological roles, such as in pathological conditions. Here, we describe a protocol to dissect human amnion tissue in three sections and maintain it in vitro. In culture, cells derived from the reflected amnion displayed a cuboidal morphology, while cells from both placental and umbilical regions were squamous. Nonetheless, all the cells obtained have an epithelial phenotype, demonstrated by the immunodetection of E-cadherin. Thus, because the placental and reflected regions in situ differ in cellular components and molecular functions, it may be necessary for in vitro studies to consider these differences, because they could have physiological implications for the use of HAEC in biomedical research and the promising application of these cells in regenerative medicine.
Introduction
Human amniotic epithelium cells (HAEC) originate during the early stages of embryonic development, at around eight days postfertilization. They arise from a population of squamous epithelial cells of the epiblast that derive from the innermost layer of the amniotic membrane1. Thus, HAEC are considered remnants of pluripotent cells from the epiblast that have the potential to differentiate into the three germ layers of the embryo2. In the last decade, diverse research groups have developed methods to isolate these cells from the amniotic membrane at the term of gestation to characterize their presumptive pluripotency-related properties in a culture model in vitro3,4.
Accordingly, it has been found that HAEC feature traits characteristic of human pluripotent stem cells (HPSC), such as the surface antigens SSEA-3, SSEA-4, TRA 1-60, TRA 1-81; the core of pluripotency transcription factors OCT4, SOX2, and NANOG; and the proliferation marker KI67, suggesting that they are self-renewing5,6,7. Moreover, these cells have been challenged using differentiation protocols to obtain cells positive for lineage-specific markers of the three germ layers (ectoderm, mesoderm, and endoderm)4,5,8, as well as in animal models of human diseases. Finally, HAEC express E-cadherin, which demonstrate that they retain an epithelial nature much like the HPSC5,9.
Apart from their embryonic origin, HAEC have other intrinsic properties that make them suitable for different clinical applications, such as the secretion of anti-inflammatory and antibacterial molecules10,11, growth factors and cytokine release12, no formation of teratomas when they are transplanted into immunodeficient mice in contrast with HPSC2, and immunological tolerance because they express HLA-G, which decreases the risk of rejection after transplantation13.
However, previous reports have assumed that the human amnion is a homogenous membrane, without considering that it can be anatomically and physiologically divided into three regions: placental (the amnion that covers the decidua basalis), umbilical (the part that envelops the umbilical cord), and reflected (the rest of the membrane not attached to the placenta)14. It has been shown that the placental and reflected regions of the amnion display differences in morphology, mitochondrial activity, detection of reactive oxygen species15, miRNA expression16, and activation of signaling pathways17. These results suggest that the human amnion is integrated by a heterogeneous population with different functionality that should be considered for further studies carried out in either in situ or in vitro models. While other laboratories have designed protocols for the isolation of HAEC from the whole membrane, our laboratory has established a protocol to isolate, culture, and characterize cells from different anatomical regions.
Protocol
This protocol was approved by the ethical committee of Instituto Nacional de Perinatología in Mexico City (Registry number 212250-21041). All procedures performed in these studies were in accordance with the ethical standards of the Instituto Nacional de Perinatología, the Helsinki Declaration, and the guidelines set forth in the Ministry of Health’s Official Mexican Standard.
1. Preparation
- Prepare a solution of 1x PBS with EDTA. To do so, add 500 μL of 0.5 M EDTA stock into 500 mL of 1x PBS for a final concentration of 0.5 mM EDTA.
- Prepare the culture medium for HAEC. Take 450 mL of high glucose-DMEM, supplement it with 5 mL of sodium pyruvate (100 mM), 50 mL of heat-inactive fetal bovine serum qualified for stem cells, 5 mL of nonessential amino acids (100x), 5 mL of antibiotic-antimycotic (100x), 5 mL of L-glutamine (200 mM), and 500 μL of mercaptoethanol (1,000x).
- Sterilize the containers for transporting and processing the tissue: a stainless steel container with a lid to transport the whole placenta from the operating theater to the laboratory, a tray (20 cm x 30 cm x 8 cm) to wash and remove blood from the whole placenta before the dissection of the amniotic membrane, and a plastic cutting board to separate the amniotic membrane into the three regions.
- Sterilize the surgical instruments (scalpels, scissors, forceps, and clamps), 500 mL beakers, cotton gauzes, and saline solution.
2. Obtaining placental tissue
NOTE: The amniotic membranes were obtained from women at full-term gestation (37−40 weeks), under indication of Cesarean delivery, without any evidence of active labor, and no microbiological characteristics of infection. The complete isolation and culture procedures were carried out within a biosecurity cabinet under sterile conditions.
- In the operating room, clamp the umbilical cord to prevent the blood flow to the rest of the tissue. Collect the entire placenta with the umbilical cord clamped in the sterile container.
- Add 500 mL of saline solution to the container to hydrate the placenta.
- Close the container and transport the tissue to the laboratory at room temperature.
- Put the container with the placenta inside the biosecurity cabinet.
NOTE: In case the dissection is not carried out within 15 min of collecting the placenta, store the container with the placenta on ice until processing. Avoid more than 1 h of elapsed time between obtaining the placenta and the start of the dissection.
3. Mechanical separation per region of the amniotic membrane
NOTE: The procedure must be carried out within a biosecurity cabinet under sterile conditions and at room temperature.
- Remove the whole placenta from the container and place it on the tray with the umbilical cord facing upward.
- Using a sterile cotton gauze, clean the blood clots from the surface of the chorion-amnion that covers the placenta.
- Identify the three regions of the membrane: the umbilical amnion enveloping the umbilical cord, the placental amnion covering the decidua basalis, and the reflected region, which is the rest of amnion that is not attached to the placenta (Figure 1).
- Dissect the umbilical amnion region (Figure 2A).
- Using dissecting forceps, hold the portion of amnion membrane that covers the junction of the placenta and umbilical cord.
- With a scalpel, dissect the region that surrounds the cord while stretching to separate it from the chorion.
- Deposit the separated tissue in a labeled beaker with 100 mL of saline solution.
- Dissect the placental amnion region (Figure 2B).
- With a sterile cotton gauze, remove the blood clots from the surface of the chorion-amnion that covers the placenta.
- Hold the membrane on the border between the placenta and the reflected region with dissecting forceps.
- Cut along the circumference of the placenta with the scalpel.
- Separate the placental amnion from the chorion, being careful not to cut any vessels from the placenta.
- Place the separated tissue in another labeled beaker with 300 mL of saline solution.
- Separate the rest of the amniotic membrane that is not attached to the placenta (i.e., the reflected portion) from the chorion (Figure 2C).
- Collect the reflected region in another labeled beaker with 300 mL of saline solution.
NOTE: Add saline solution continually during the dissection to prevent tissue from drying out.
- Collect the reflected region in another labeled beaker with 300 mL of saline solution.
4. Washing the membranes
NOTE: The procedure must be carried out within a biosecurity cabinet under sterile conditions at room temperature.
- Discard the saline solution of each membrane region separately.
- Add 100 mL of fresh saline solution to the umbilical region.
- Add 300 mL of fresh saline solution to the placental and reflected regions respectively.
- Stir the membranes with the help of dissecting forceps to remove blood residue.
- Discard the saline solution.
- Repeat the washes and agitation at least 3x until the membranes are translucent.
- Place and extend the membranes on the board to clean with sterile gauze the blood clots that were not removed with the washes.
NOTE: It is very important to remove as many erythrocytes as possible, as their presence affects the trypsin function and the viability of subsequent cell cultures.
5. Enzymatic digestion of the membranes from different regions
NOTE: The procedure must be carried out within a biosecurity cabinet under sterile conditions.
- Cut the reflected and placental regions into two or three fragments.
- Do not cut the umbilical region.
- Place the fragments of each region in centrifuge tubes. Add 20 mL of 0.5% trypsin/EDTA to the reflected and placental regions and 5 mL of 0.5% trypsin/EDTA to the umbilical region, respectively.
NOTE: It is important to cut the reflected and placental regions into smaller pieces because they need to be completely immersed in the trypsin solution. - Shake the centrifuge tubes lightly for 30 s. Discard the trypsin.
- Add 30 mL of new 0.5% trypsin/EDTA to the reflected and placental regions and 15 mL of new 0.5% trypsin/EDTA to the umbilical region, respectively.
- Place the tubes in a rotator inside the incubator.
- Incubate with rotation (20 rpm) for 40 min at 37 °C.
NOTE: If a tube rotator is not available, shake the tubes lightly manually every 10 min. - Transfer the trypsin/cell solutions from each region into new centrifuge tubes.
- Add 2x the volume of HAEC media prewarmed at 37 °C per tube to inactivate the enzyme.
- Store the first digestion on ice.
- Repeat steps 5.6−5.8 for a second digestion period.
- For each region, hold one end of the amnion portion using dissecting forceps, and with another pair squeeze along the tissue to remove rows of epithelial cells that did not completely peel off during previous incubation periods.
- Collect the second digestion into another set of centrifuge tubes and inactivate with 2x the volume of HAEC media.
- Discard the digested membranes into a biohazard container.
6. Isolation of the HAEC
NOTE: The procedure must be carried out within a biosecurity cabinet under sterile conditions.
- Centrifuge all tubes at 200 x g for 10 min at 4 °C.
- Discard the supernatant and add 10 mL of prewarmed HAEC media (to 37 °C) per tube and pipet gently to disaggregate each pellet.
- Combine the cellular suspensions of the two digestions in an individual tube for each membrane region.
- Filter the cellular suspensions using 100 μm cell strainers to remove the extracellular matrix debris and obtain single cells.
- Prepare three aliquots with 90 µL of trypan blue in a microcentrifuge tube.
- Add 10 μL of each cell suspension per membrane region into the microcentrifuge tubes and mix.
- Count the cells with a hemocytometer under a light field microscope.
7. Culture of HAEC
- Seed the HAEC from the three regions separately at a density of 3 × 104 cells/cm2 with prewarmed HAEC media, supplemented with 10 ng/mL of human epidermal growth factor (EGF).
- Seed the cells in 100 mm plates to maintain them in vitro or in 24 well plates for immunochemical analysis.
- Incubate the dishes at 37 °C under normoxic conditions (5% CO2) in a humidified incubator.
- Add EGF daily and change the medium every third day.
NOTE: The cells will become confluent after 4−6 days. - Use the cells for conventional immunohistochemistry, cell sorting analysis, cryopreserving, RNA and protein extraction, or to continue the passage.
8. Passage of HAEC
- Remove the HAEC medium and wash 2x with PBS/EDTA 0.01M solution.
- Incubate with a PBS/EDTA 0.01M solution for 15 min at 37 °C.
- Remove the PBS/EDTA and add 1.5 mL of 0.5% trypsin/EDTA.
- Incubate for 5−8 min at 37 °C.
- Inactivate the enzyme with two volumes of HAEC media.
- Collect the mixed solution and centrifuge for 5 min at 200 x g.
- Count the cells and continue with following passages or use the cells for further analysis.
Representative Results
HAEC were isolated from each of the three anatomical regions of the amniotic membrane and individually cultured in vitro. After 48 h of culture, cells with an epithelial phenotype adhered to the surface of the plate, although the media also contained cell debris and floating cells, which were removed once the medium was changed (Figure 3).
During the processing of primary culture (passage zero, P0), some complications could arise that can interfere with the experimental data analysis (Figure 4): it is advisable to discard the cultures and process another membrane upon identifying the presence of bacteria due to contamination of the reagents or during the isolation process (Figure 4A); excessive erythrocytes due to insufficient washing of the membranes (Figure 4B); deficient or no adhesion of the cells to the plates (Figure 4C); or cells with fibroblast morphology (Figure 4D), suggesting that human amniotic mesenchymal cells (HAMC) were isolated instead of HAEC.
HAEC morphology depends on the origin of these cells: cells from the reflected zone have a cuboidal morphology and grow in a cobbled monolayer, unlike cells from the placental and umbilical regions, which are flatter and squamous (Figure 5). These data support that the epithelial layer from the amnion is not uniform throughout the membrane. During the passages, the size of the cells from all regions increases, but they maintain their epithelial nature and do not acquire a fibroblast morphology. Indeed, immunofluorescence against E-cadherin showed that the primary cultures (P0) and subcultures (P1-P2) maintained their epithelial phenotype (Figure 6), suggesting that there is no contamination of another cell type, such as mesenchymal stromal cells, or epithelial-mesenchymal transition. In addition, these cells show no evidence of cell death according to the TUNEL assay (Supplementary Figure 1) but are positive for the KI-67 proliferation marker (Figure 6), although our previous results found no significant differences for this marker between each passage5.
The number of cells obtained varies according to the region: 61.6 x 106 and 71.8 x 106 cells from the reflected and placental regions, respectively, and less than 1 x 106 per sample from the umbilical region (Table 1). It has been reported with this protocol that placental and umbilical region are very similar in their expression profiles, as opposed to the reflected and placental regions, especially in genes that participate in ECM receptor interaction, focal adhesion, and the PI3K-Akt signaling pathway through RNA-seq6. In agreement, a previous study reported the differential expression of mitogen-activated protein kinase and transforming growth factor beta pathways, as well as proinflammatory cytokines between both regions17.
Although the evidence showed that the subpopulations from the amnion differ in their morphology and physiological properties, we previously demonstrated that the expression and presence of the core of the pluripotency factors does not change in HAEC derived from placental and reflected regions6. In this context, in addition to the relatively limited number of cells from the umbilical region, subsequent studies should focus on isolated HAEC from the placental and reflected amnion independently, considering the physiological implications, because the different subpopulations would not respond equally to specific events, such as prolonged pregnancy or inflammatory processes during labor, although there are no specific positive cells to the main pluripotency markers (Figure 7).
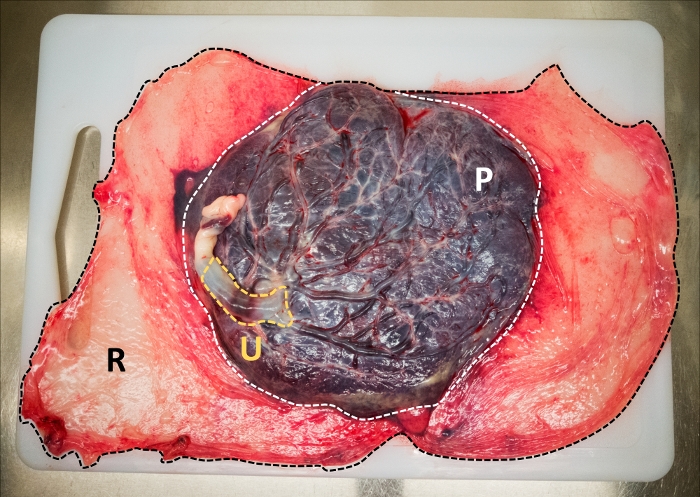
Figure 1: Anatomical regions from the amniotic membrane at term. Umbilical amniotic membrane (yellow), placental amniotic membrane (white), and reflected amniotic membrane (black). Please click here to view a larger version of this figure.
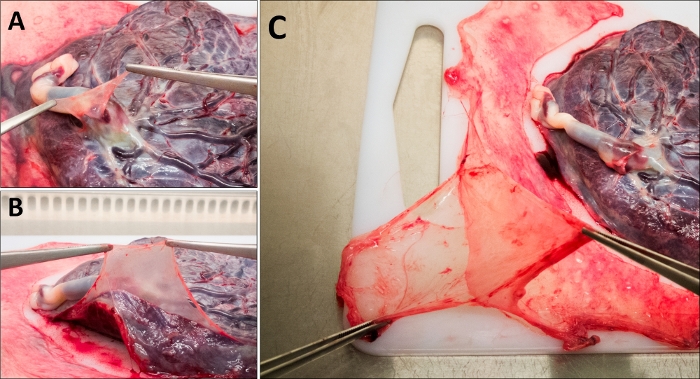
Figure 2: Mechanical dissection of the amniotic membrane. (A) Umbilical region, (B) placental region, and (C) reflected region. Please click here to view a larger version of this figure.
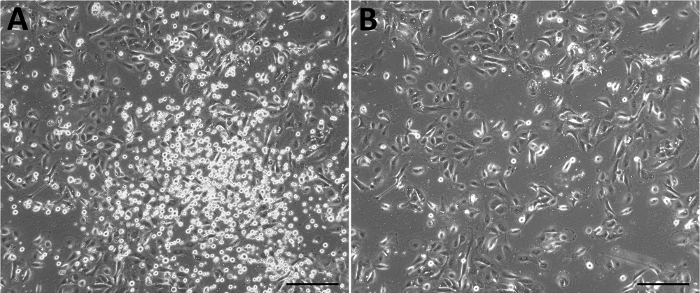
Figure 3: Representative primary culture of HAEC derived from the amniotic membrane. Culture 48 h after isolation without (A) and with (B) a change of media. Scale bars = 50 µm. Please click here to view a larger version of this figure.
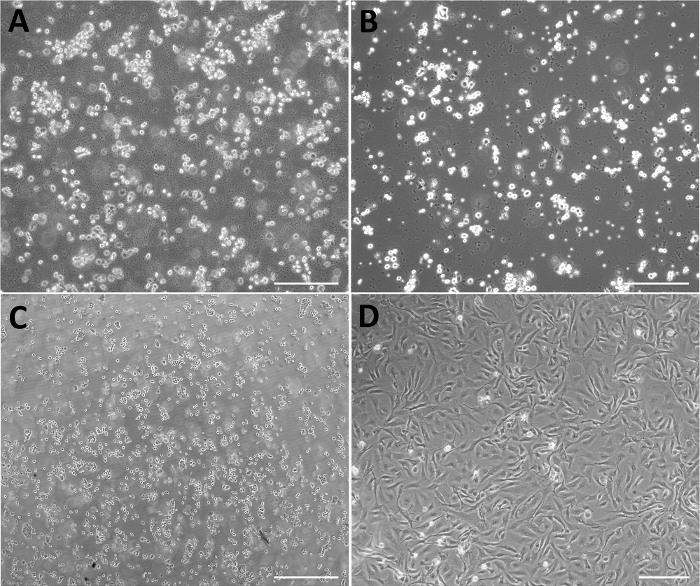
Figure 4: Representative micrographs of negative results of primary culture of HAEC. (A) Excess of erythrocytes. (B) Bacterial contamination. (C) HAEC not adhered after 48 h of isolation. (D) Primary culture composed of cells with fibroblast morphology. Scale bars = 200 µm. Please click here to view a larger version of this figure.
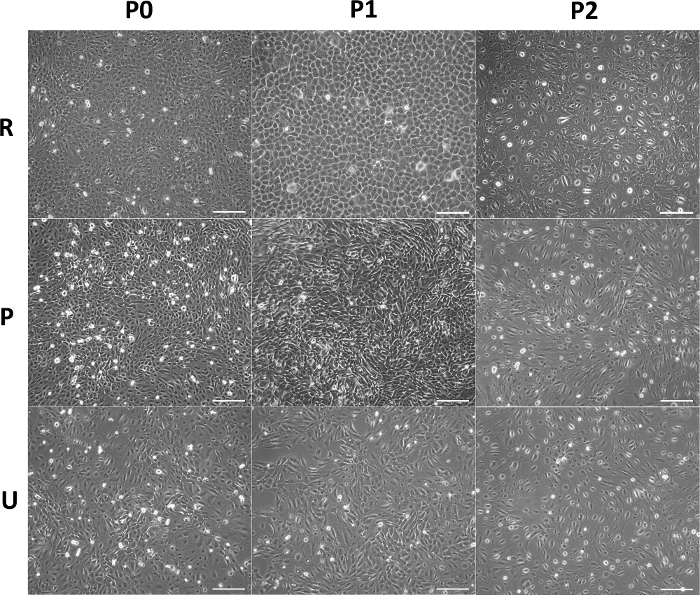
Figure 5: Morphology of HAEC from different anatomical regions in vitro. Representative micrographs of confluent HAEC from reflected (upper panel), placental (middle panel), and umbilical (lower panel) regions cultured though P0−P2. Scale bars 200 = µm. Please click here to view a larger version of this figure.
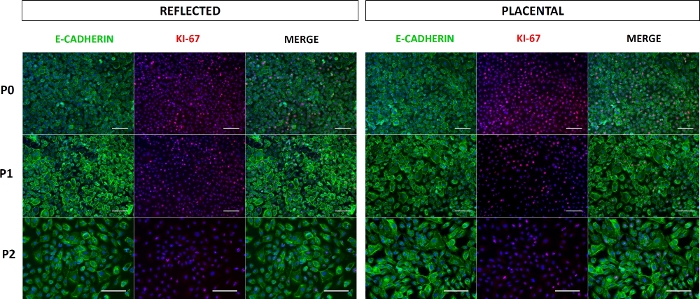
Figure 6: Expression of E-cadherin and KI-67 in HAEC. Representative epifluorescence microscopy images of E-cadherin+ and KI-67+ HAEC from reflected (left panel) and placental (right panel) amnion cultured through P0−P2. Scale bars = 100 µm. Please click here to view a larger version of this figure.
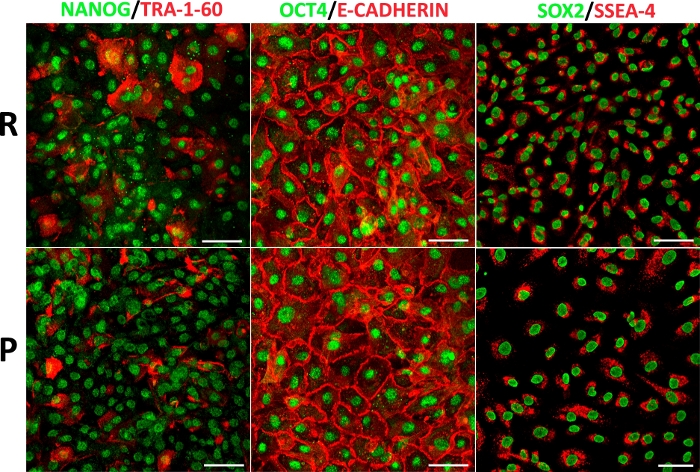
Figure 7: HAEC from different anatomical regions display a pluripotency marker panel. Representative confocal microscopy images of double immunofluorescence amnion for NANOG with TRA-1-60, OCT4 with E-cadherin, and SOX2 with SSEA-4 in HAEC (P1) from reflected (upper panel) and placental (lower panel) regions. Scale bars = 100 µm. Please click here to view a larger version of this figure.
| # MEMBRANE | REFLECTED | PLACENTAL | UMBILICAL | COMPLETE MEMBRANE |
| 1 | 75 X 106 | 130 X 106 | 0.2 X 106 | 205.2 X 106 |
| 2 | 38.5 X 106 | 52 X 106 | 0.53 X 106 | 91.03 X 106 |
| 3 | 59 X 106 | 53 X 106 | 0.2 X 106 | 112.2 X 106 |
| 4 | 42 X 106 | 27 X 106 | 0.36 X 106 | 69.36 X 106 |
| 5 | 44.8 X 106 | 22.3 X 106 | NP | 76.1 X 106 |
| 6 | 100 X 106 | 140 X 106 | 1 X 106 | 241 X 106 |
| 7 | 72 X 106 | 78 X 106 | NP | 150 X 106 |
| AVERAGE | 61.6 X 106 | 71.8 X 106 | 0.45 X 106 | 133.8 X 106 |
Table 1: Number of HAEC isolated per region from amniotic membranes. (NP = not processed).
Supplementary Figure 1: TUNEL staining in HAEC (passage 2) from reflected region and in positive control cells (HL-60 cells treated with camptothecin). Scale bars = 50 µm. Please click here to download this figure.
Discussion
We implemented a new protocol to isolate HAEC from term membranes. It differs from previous reports in that each membrane was divided into its three anatomical regions prior to isolation to analyze cells from each one.
One of the most critical steps in the protocol is the washing of the membrane to remove all blood clots, because they can interfere with the activity of trypsin when separating the epithelial cells. Failure to carry out this step properly can lead to obtaining a primary culture with excessive erythrocytes and few adherent epithelial cells. If no attachment is observed in the first 24−48 h after seeding the cells, we recommend discarding the culture. Another critical point is the incubation time for the enzymatic digestion. If the incubation period is too long, cell viability may decrease and/or cell cultures with mesenchymal instead of epithelial morphology may be obtained (Figure 4). Also, if membrane digestion is not done with the proper agitation, the probability of obtaining a low number of cells per membrane increases.
In this protocol, we can maintain populations of HAEC in vitro without losing their epithelial phenotype, as demonstrated by the presence of specific epithelial cell marker E-cadherin for most cells along the passages (Figure 6). However, in our experience the HAEC can be only be expanded for three passages (P1−P3). The limited number of passages should be taken into consideration for experiments that require long periods of culture or multiple passages. In addition, it has been reported that after P3 the cells begin to change their morphology and reduce the expression of E-cadherin, so the prolonged culture could induce epithelial-mesenchymal transition (EMT)18. Recently, it was demonstrated that progesterone prevents EMT in ovine amniotic epithelial cells19,20, so it would be interesting to analyze the effect of different concentrations of progesterone in the HAEC culture medium to avoid the mesenchymal phenotype after the third passage.
In all previous reports where HAEC were isolated, the amnion has been assumed to be a uniform tissue despite the possible heterogeneity of epithelial populations that comprise the whole membrane. In contrast, this protocol divides the membrane into its three anatomical areas to separately isolate and culture HAEC considering the previous morphological and gene expression reports that suggest functional differences. It is also unnecessary to purify through cell sorting to obtain only epithelial populations, as demonstrated by detection of the E-cadherin marker during passages until P2.
It has been shown that isolated HAEC from each of the membrane regions have a similar expression of pluripotency core, being a latent reservoir of cells with stemness. In this context, these cells can be used in vitro to study molecular mechanisms, such as the dynamic localization of pluripotency-related transcription factors or their ability to remain quiescent despite expressing cancer-associated genes, regardless of their anatomical region of origin. However, future research must consider molecular differences reported (global expression genes, metabolic activity, signaling pathways) between each region, which would have repercussions for their application in regenerative medicine. We previously reported a different expression of genes involved in PI3K/AKT and focal adhesion signaling pathways between the different amniotic regions by RNA-seq6. PI3K/AKT regulates self-renewal and maintains pluripotency in HPSC, while activation of focal adhesion kinase promotes their differentiation21,22. Thus, we propose to characterize the role of both pathways in HAEC isolated from different anatomical regions during lineage-specific differentiation protocols. In addition, several studies demonstrate the differences between both regions regarding cellular components, molecular function, and biological processes. For example, the region-specific gene expression and protein distribution of aquaporins, which are involved in the transport process of intramembranous absorption, have been reported23. Another study compared the miRNome by microarrays, showing region-specific expression of miR-145 and miR-143, the latter being able to posttranscriptionally regulate the prostaglandin-endoperoxidase synthase 2 under specific conditions such as labor at term16. Moreover, the placental region presents higher mitochondrial respiration but lower reactive oxygen species detection as compared with reflected tissue15. Dissecting the amnion in reflected and placental regions is encouraged to isolate HAEC and analyze their peculiar properties separately, such as the release of growth factors and cytokines, their low immunogenicity, production of extracellular matrix, the effect of their conditioned medium in coculture with other cell types, and novel functions such as their capacity to maintain HPSC lines as a feeder layer24.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
Our research was supported by grants from Instituto Nacional de Perinatología de México (21041 and 21081) and CONACYT (A1-S-8450 and 252756). We thank Jessica González Norris and Lidia Yuriria Paredes Vivas for the technical support.
Materials
| Culture reagents | |||
| 2-Mercaptoethanol | Thermo Fisher Scientific/Gibco | 21985023 | 55 mM |
| Animal-Free Recombinant Human EGF | Peprotech | AF-100-15 | |
| Antibiotic-Antimycotic | Thermo Fisher Scientific/Gibco | 15240062 | 100X |
| Dulbecco's Modified Eagle Medium | Thermo Fisher Scientific/Gibco | 12430054 | Supplemented with high glucose and HEPES |
| EDTA | Thermo Fisher Scientific/Ambion | AM9260G | 0.5 M |
| Embryonic stem-cell FBS, qualified | Thermo Fisher Scientific/Gibco | 10439024 | |
| Non-Essential Amino Acids | Thermo Fisher Scientific/Gibco | 11140050 | 100X |
| Paraformaldehyde | any brand | ||
| Phosphate-Buffered Saline | Thermo Fisher Scientific/Gibco | 10010023 | 1X |
| Saline solution (sodium chloride 0.9%) | any brand | ||
| Sodium Pyruvate | Thermo Fisher Scientific/Gibco | 11360070 | 100 mM |
| Trypsin/EDTA 0.05% | Thermo Fisher Scientific/Gibco | 25300054 | |
| Disposable material | |||
| 100 µm Cell Strainer | Corning/Falcon | 352360 | |
| 100 mm TC-Treated Culture Dish | Corning | 430167 | |
| 24-well Clear TC-treated Multiple Well Plates | Corning/Costar | 3526 | |
| 6-well Clear TC-treated Multiple Well Plates | Corning/Costar | 3516 | |
| Non-Pyrogenic Sterile Centrifuge Tube | any brand | with conical bottom | |
| Non-Pyrogenic sterile tips of 1,000 µl, 200 µl and 10 µl. | |||
| Sterile cotton gauzes | |||
| Sterile serological pipettes of 5, 10 and 25 mL | any brand | ||
| Sterile surgical gloves | any brand | ||
| Equipment | |||
| Biological safety cabinet | |||
| Centrifuge | |||
| Micropipettes | |||
| Motorized Pipet Filler/Dispenser | |||
| Sterile beakers of 500 mL | |||
| Sterile plastic cutting board | |||
| Sterile scalpels, scissors, forceps, clamps | |||
| Sterile stainless steel container | |||
| Sterile tray | |||
| Tube Rotator | MaCSmix | ||
| Antibodies and Kits | Antibody ID | ||
| Anti-E-cadherin | BD Biosciences | 610181 | RRID:AB_3975 |
| Anti-KI67 | Santa Cruz | 23900 | RRID:AB_627859) |
| Anti-NANOG | Peprotech | 500-P236 | RRID:AB_1268274 |
| Anti-OCT4 | Abcam | ab19857 | RRID:AB_44517 |
| Anti-SOX2 | Millipore | AB5603 | RRID:AB_2286686 |
| Anti-SSEA-4 | Cell Signaling | 4755 | RRID:AB_1264259 |
| Anti-TRA-1-60 | Cell Signaling | 4746 | RRID:AB_2119059 |
| Goat Anti-Mouse Alexa Fluor 488 | Thermo Fisher Scientific | A-11029 | RRID:AB_2534088 |
| Goat Anti-Rabbit Alexa Fluor 568 | Thermo Fisher Scientific | A-11036 | RRID:AB_10563566 |
| Tunel Assay Kit | Abcam | 66110 |
Referencias
- Shahbazi, M. N., et al. Self-organization of the human embryo in the absence of maternal tissues. Nature Cell Biology. 18 (6), 700-708 (2016).
- Garcia-Lopez, G., et al. Human amniotic epithelium (HAE) as a possible source of stem cells (SC). Gaceta Medica de Mexico. 151 (1), 66-74 (2015).
- Gramignoli, R., Srinivasan, R. C., Kannisto, K., Strom, S. C. Isolation of Human Amnion Epithelial Cells According to Current Good Manufacturing Procedures. Current Protocols in Stem Cell Biology. 37, (2016).
- Murphy, S., et al. Amnion epithelial cell isolation and characterization for clinical use. Current Protocols in Stem Cell Biology. , (2010).
- Garcia-Castro, I. L., et al. Markers of Pluripotency in Human Amniotic Epithelial Cells and Their Differentiation to Progenitor of Cortical Neurons. PLoS One. 10 (12), 0146082 (2015).
- Garcia-Lopez, G., et al. Pluripotency markers in tissue and cultivated cells in vitro of different regions of human amniotic epithelium. Experimental Cell Research. 375 (1), 31-41 (2019).
- Yang, P. J., et al. Biological characterization of human amniotic epithelial cells in a serum-free system and their safety evaluation. Acta Pharmacological Sinica. 39 (8), 1305-1316 (2018).
- Zou, G., et al. MicroRNA32 silences WWP2 expression to maintain the pluripotency of human amniotic epithelial stem cells and beta isletlike cell differentiation. International Journal of Molecular Medicine. 41 (4), 1983-1991 (2018).
- Avila-Gonzalez, D., et al. Capturing the ephemeral human pluripotent state. Developmental Dynamics. 245 (7), 762-773 (2016).
- Niknejad, H., et al. Properties of the amniotic membrane for potential use in tissue engineering. European Cells & Materials. 15, 88-99 (2008).
- Miki, T. Stem cell characteristics and the therapeutic potential of amniotic epithelial cells. American Journal of Reproductive Immunology. 80 (4), 13003 (2018).
- Wu, Q., et al. Comparison of the proliferation, migration and angiogenic properties of human amniotic epithelial and mesenchymal stem cells and their effects on endothelial cells. International Journal of Molecular Medicine. 39 (4), 918-926 (2017).
- Hammer, A., et al. Amnion epithelial cells, in contrast to trophoblast cells, express all classical HLA class I molecules together with HLA-G. American Journal of Reproductive Immunology. 37 (2), 161-171 (1997).
- Benirschke, K., et al. Anatomy and Pathology of the Placental Membranes. Pathology of the Human Placenta. , 268-318 (1995).
- Banerjee, A., et al. Different metabolic activity in placental and reflected regions of the human amniotic membrane. Placenta. 36 (11), 1329-1332 (2015).
- Kim, S. Y., et al. miR-143 regulation of prostaglandin-endoperoxidase synthase 2 in the amnion: implications for human parturition at term. PLoS One. 6 (9), 24131 (2011).
- Han, Y. M., et al. Region-specific gene expression profiling: novel evidence for biological heterogeneity of the human amnion. Biology of Reproduction. 79 (5), 954-961 (2008).
- Alcaraz, A., et al. Autocrine TGF-beta induces epithelial to mesenchymal transition in human amniotic epithelial cells. Cell Transplantation. 22 (8), 1351-1367 (2013).
- Canciello, A., et al. Progesterone prevents epithelial-mesenchymal transition of ovine amniotic epithelial cells and enhances their immunomodulatory properties. Scientific Reports. 7 (1), 3761 (2017).
- Canciello, A., Greco, L., Russo, V., Barboni, B. Amniotic Epithelial Cell Culture. Methods in Molecular Biology. 1817, 67-78 (2018).
- Singh, A. M., et al. Signaling network crosstalk in human pluripotent cells: a Smad2/3-regulated switch that controls the balance between self-renewal and differentiation. Cell Stem Cell. 10 (3), 312-326 (2012).
- Villa-Diaz, L. G., Kim, J. K., Laperle, A., Palecek, S. P., Krebsbach, P. H. Inhibition of Focal Adhesion Kinase Signaling by Integrin alpha6beta1 Supports Human Pluripotent Stem Cell Self-Renewal. Stem Cells. 34 (7), 1753-1764 (2016).
- Bednar, A. D., Beardall, M. K., Brace, R. A., Cheung, C. Y. Differential expression and regional distribution of aquaporins in amnion of normal and gestational diabetic pregnancies. Physiological Reports. 3 (3), 12320 (2015).
- Avila-Gonzalez, D., et al. Human amniotic epithelial cells as feeder layer to derive and maintain human embryonic stem cells from poor-quality embryos. Stem Cell Research. 15 (2), 322-324 (2015).

