Experimental Analysis of Apoptotic Thymocyte Engulfment by Macrophages
Summary
Here, we present a protocol to prepare apoptotic thymocytes and peritoneal macrophages and analyze the efficiency of efferocytosis and the specific inhibitor-mediated blocking of apoptotic thymocytes engulfment. This protocol has a broad application in cell-mediated clearance of other particles including artificial beads and bacteria.
Abstract
Cell apoptosis is a natural process and plays a critical role in embryonic development, homeostatic regulation, immune tolerance induction, and resolution of inflammation. Accumulation of apoptotic debris in the body may trigger chronic inflammatory responses that lead to systemic autoimmune diseases over time. Impaired apoptotic cell clearance has been implicated in a variety of autoimmune diseases. Apoptotic clearance is a complex process rarely detected under physiological conditions. It involves abundant surface receptors and signaling molecules. Studying the process of apoptotic cell clearance provides insightful molecular mechanisms and subsequent biological responses, which may lead to the development of new therapeutics. Here, we describe protocols for the induction of apoptotic thymocytes, the preparation of peritoneal macrophages, and the analysis of apoptotic cell clearance by flow cytometry and microscopy. All cells will undergo apoptosis at a certain stage, and many residential and circulating cells can uptake apoptotic debris. Therefore, the protocol described here can be used in many applications to characterize apoptotic cell binding and ingestion by many other cell types.
Introduction
Our body generates 1-10 billion apoptotic cells on a daily basis. Such a large number of apoptotic cells must be cleared in a way that the immune responses remain quiescent. To ensure the clearance of apoptotic cells in a timely manner, numerous types of tissue resident cells and circulating cells develop mechanisms to engulf apoptotic cells1. Dysfunctional regulation of apoptosis has been implicated in the onset and progression of various inflammatory disease and autoimmunity2. Apoptosis also plays a critical role in the pathogenesis of cancer development and its subsequent resistance to conventional treatments3,4. Removal of apoptotic cells generally promotes an anti-inflammatory response, which may be linked to immunological tolerance5. Disturbance of apoptotic cell clearance drives self-immunization and contributes to the development of systemic autoimmune diseases in both humans and mice6.
When cells undergo apoptosis, they expose the phosphatidylserine (PtdSer) from the inner leaflet to the outer leaflet of the membrane. PtdSer will then be recognized by phagocytes through surface receptors. Over a dozen receptors have been identified to recognize and/or facilitate the engulfment of apoptotic cells. In general, there are at least three types of surface receptors involved in the apoptotic cell clearance: tethering receptors, recognize apoptotic cells; tickling receptors, initiate engulfment; chaperoning receptors, facilitate the whole process7. TAM receptor tyrosine kinases (TAM RTKs) consist of Tyro-3, Axl, and Mer and are primarily expressed by myeloid cells of the immune system8. The primary function of TAM RTKs is to serve as tethering receptors, facilitating the phagocytic removal of apoptotic cells and debris. Our group has studied TAM mediated apoptotic cell clearance in the setting of autoimmunity for many years. The vitamin K-dependent protein growth arrest specific protein 6 (Gas6) and protein S (ProS) binds to and activates TAM receptors9,10. Gas6 is produced in the heart, kidneys, and lungs. ProS is mainly produced in the liver11. TAM recognizes of apoptotic cells in such a way that the N-terminal of Gas6/ProS binds to the PtdSer on an apoptotic cell and the C-terminal of Gas6/ProS binds to TAM receptors that anchored on the surface of phagocytes. Together with the other receptors, engulfment of apoptotic cells occurs12. Though Mer can bind to both the ligands ProS and Gas6, we found that Gas6 appears to be the sole ligand for Mer-mediated macrophage phagocytosis of apoptotic cells, which can be blocked by anti-Mer antibody13. Macrophages are professional phagocytes. Rapid clearance of apoptotic cells by macrophages is important for the inhibition of inflammation and autoimmune responses against intracellular antigens. Mer receptor tyrosine kinase is critical for the macrophage engulfment and efficient clearance of apoptotic cells14. In mouse spleen, Mer mainly expresses on the marginal zone and tangible body macrophages13.
The protocol presented here describes a basic method to induce cell apoptosis and demonstrate ways to measure the process and the efficiency of efferocytosis. These protocols can be readily adapted to study efferocytosis by other cell types in engulfment of apoptotic cells of different origins.
Protocol
Experimental mice were bred and maintained in our mice colony. All animal work was conducted according to the guidelines of the Institutional Animal Care and Use Committee (IACUC) of the University of Cincinnati.
1. Preparation of CFSE labeled apoptotic thymocytes
- Euthanize two naïve C57/B6 mice by CO2 inhalation for 10 min and dissect to open the chest cavity, remove (pull out) the thymus with curved fine-tip forceps into tissue culture petri dish containing 10 mL of RPMI1640 medium.
- Obtain single cell suspension by grinding the whole thymus against two frosted ends of the microscope slides and then filter the suspension through 100 μm cell strainer.
- Collect the 10 mL thymus suspension into a 50 mL tube and centrifuge at 300 x g for 5 min.
- Remove the supernatant, resuspend in 40 mL of 1x PBS, and count cell numbers with a hemocytometer.
- Centrifuge at 300 x g for 5 min and remove the supernatant.
- Resuspend in 20 mL of 1x PBS in a 50 mL tube as a single cell suspension with up to 2 x 108 cells (if more than 2 x 108 cells, resuspend the rest of the cells in another 20 mL of 1 x PBS in a different 50 mL tube).
- Make 5 μM of CFSE in equal volume (20 mL) of 1x PBS in a separate 50 mL tube by pipetting 40 μL of CFSE stock solution (2.5 mM) into 20 mL of 1x PBS and mix well by inverting the tube 2 – 3 times.
- Add the 20 mL of CFSE from step 1.7 into the 20 mL of cell suspension from step 1.6 (the final concentration of CFSE is now 2.5 μM).
NOTE: For every 20 mL cell suspension, a 20 mL of CFSE tube is needed. - Invert the cell and CFSE mixture tube 2 – 3 times and incubate the mixture in the dark at room temperature for a maximum of 2 min, then stop the reaction by adding 10 mL of heat-inactivated horse serum.
- Centrifuge the 50 mL tube mixture at 300 x g for 5 min at room temperature.
NOTE: If the CFSE labeling is successful, cell pellet will become light yellow in color. - Remove the supernatant and resuspend the cell pellet in 40 mL of 1x PBS and count cell numbers with a hemocytometer.
- Centrifuge the cell suspension at 300 x g for 5 min and discard the supernatant.
- Wash the cell pellet again with 40 mL of RPMI1640 medium.
- Remove the supernatant and resuspend cells with RPMI1640 tissue culture medium (RPMI1640, 20 mM HEPES, 10% FBS (heat inactivated), 20 mM glutamine, and 1x Pen/Strep) at a concentration of 7 x 106 cells/mL in a 100 mm tissue culture dish.
NOTE: If more cells are obtained, a separate 100 mm tissue culture dish will be needed. - Add staurosporine into the cell suspension culture at a final concentration of 1 μM and culture for 4 h at 37 °C in a tissue culture incubator supplied with 5% CO2.
2. Preparation of peritoneal macrophages
- Inject two C57B6 mice (or any gene-manipulated mice in the lab) intraperitoneally with 1 mL of 3% aged thioglycollate at day 0.
- Euthanize the mice at day 5 as in step 1.1, cut and peel open the abdominal skin but leave the peritoneum intact. Flush the peritoneal cavity by quickly pushing 10 mL of wash buffer (RPMI1640, 2% FBS, 0.04% EDTA) into the peritoneal cavity using a 10 mL syringe attached with a 18 G needle.
- Retrieve the wash buffer slowly with the same needle/syringe, and collect the wash buffer into a 50 mL tube (details refer to Janssen lab article15).
- Wash the peritoneal gavage twice with 1x PBS, resuspend the peritoneal macrophages in RPMI1640 tissue culture medium at a density of 2 x 106 cells/mL and aliquot 500 μL into each well of the 24-well plate. Leave the plate in a tissue culture incubator for 2 h.
- Remove the floating cells by aspirating and replacing with 500 μL of fresh culture medium, twice.
- Optionally, add TAM receptor tyrosine inhibitor, RXDX-106, at concentrations indicated in the figure legends into each well of the macrophage culture and incubate for another 2 h.
3. Co-culture of peritoneal macrophages with apoptotic thymocytes
- Collect apoptotic thymocytes from step 1 and wash three times with RPMI1640 medium. The efficiency of apoptotic induction can be measured at this stage with the Annexin V/7-AAD kit.
- Distribute 0 – 12 x 106 cells (in 500 μL medium) into each well of the macrophage cultures from protocol #2, according to the experimental arrangement (e.g., see Figure 1). This makes the whole culture volume of 1 mL in each well of the 24-well plate. Add the blocking antibody into the culture immediately before the addition of apoptotic thymocytes.
- Culture the cell mixture at 37 °C for 4 h in a tissue culture incubator supplied with 5% CO2.
- Wash each well of the culture with 1x PBS (containing 500 μM EDTA) twice to remove the free-floating apoptotic cells.
- Stain the plate-bound macrophages with CD11b-PE at this stage.
- Wash the plate-bound macrophages with staining buffer (1x PBS, 1% BSA) once.
- Add 200 μL of staining buffer containing 2 μL CD11b-PE into each well.
- Incubate the plate at 4 °C for 20 min.
- Wash each well of plate three times with the staining buffer.
- Add 200 μL of staining buffer and proceed the plate for image analysis under the fluorescent microscope.
- Alternatively, detach the plate-bound macrophages by adding 1 mL of 1% lidocaine in PBS and incubating for 10 min at 37 °C.
- Detach plate-bound macrophages with repeat pipetting.
- Transfer macrophage suspension into an individual 5 mL round-bottom FACS tube from each well of the 24-well plate.
- Centrifuge at 300 x g for 5 min.
- Remove the supernatant and add 200 μL of staining buffer containing 2 μL of CD11b-PE (1:200 dilution in staining buffer).
- Incubate the cell suspension in staining buffer at 4 °C for 20 min.
- Wash the cell suspension twice with staining buffer.
- Add 200 μL of staining buffer and proceed for FACS analysis with a flow cytometer and analyze for the percentage of CFSE positivity macrophages.
Representative Results
Analysis of peritoneal macrophage-mediated engulfment of apoptotic thymocytes. Peritoneal macrophages and apoptotic cells were prepared and co-cultured as described in the protocol. Macrophages were detached and stained with PE conjugated anti-CD11b antibody for 20 min on ice. Macrophages were then washed and processed in a flow cytometer. As seen, there is no CFSE positive macrophage in the bottom right quadrant when no apoptotic cells were added into the culture (Figure 1A and Figure 2, first panel). Thioglycollate-stimulated peritoneal macrophages have the variable capacity to engulf apoptotic cells. Up to 30% of macrophages showed positive in the CFSE channel, indicating they have ingested CFSE-labeled apoptotic cells in this experiment (Figure 1). It is worth to note that CFSE positive macrophages spread out in the bottom right quadrant due to different intensities, indicating that the number of apoptotic cells within macrophages is different. Therefore, microscopic observation of macrophage engulfment of apoptotic cells is essential to investigate the capacity of macrophages to ingest apoptotic cells (Figure 2). The higher ratio of apoptotic cells to macrophages not only increases the number of macrophages ingesting apoptotic cells but also enhances the ability of macrophages to ingest more apoptotic cells (Figure 2).
Dose-dependent inhibition of efferocytosis by Mer blockage.
In another set of experiments, we found that about 15% of the macrophages became CFSE positive when CFSE-labeled apoptotic thymocytes (ration of 6:1) were added into the macrophage culture for 4 h, indicating they are the phagocytic macrophages (Figure 3B). One function of Mer on macrophages is to recognize and mediate phagocytosis of apoptotic cells through the bridging molecule, Gas6. To test the percentage of efferocytosis attributed by Mer inhibition, we added anti-Mer antibodies into the culture to block Mer-mediated efferocytosis. Mer antibody blocks macrophage efferocytosis in a dose-dependent manner (Figure 3C, Figure 3D) and the overall blockage may account for about 30% of the efferocytosis efficiency in the current setting (Figure 3), This data was also confirmed in our previous study with Mer knockout macrophages13. We then tested the efficiency of inhibition with the newly FDA approved TAM receptor inhibitor, RXDX-106, which inhibits all TAM receptors with different affinities (Axl>>Tyro3>Mer). RXDX-106 was added into the macrophage culture 2 hours before co-incubation with apoptotic thymocytes. As shown in Figure 4, RXDX-106 inhibited macrophage efferocytosis in a dose dependent manner. The saturated inhibition concentration was about 100 nM (Figure 4, solid line), a concentration that appears to be more effective than Mer-deficiency (Figure 4 dotted line) or Mer antibody (Figure 3, panel D) alone. Since macrophage expresses all three TAM receptors on the surface16, it is expected to see that higher concentrations of RXDX-106 (over 100 nM) will block all three receptors, and therefore, will be more effective in blocking efferocytosis than targeting a single TAM receptor, Mer.
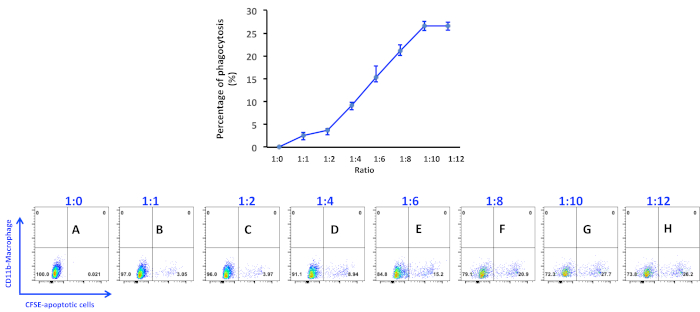
Figure 1. Percentage of phagocytosis of apoptotic thymocytes by peritoneal macrophages. Apoptotic thymocytes were induced by incubating with 1 μM of staurosporine for 4 h and added into macrophage culture at a ratio as indicated in the figure panels: (A) 1:0; (B) 1:1; (C) 1:2; (D) 1:4; (E) 1:6; (F) 1:8; (G) 1:10; (H) 1:12. Data were acquired with a flow cytometer. Percentage of phagocytic macrophages was analyzed using the software associated with the flow cytometer. Please click here to view a larger version of this figure.
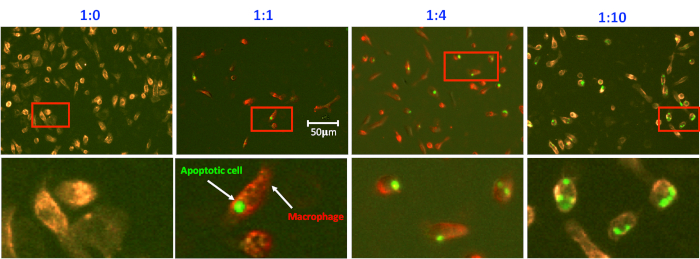
Figure 2. Microscopic analysis of efferocytosis. Efferocytosis was prepared as in Figure 1. Phagocytic macrophages were stained in situ on the plate with CD11b-PE, fixed with paraformaldehyde, and evaluated. Images were acquired using the fluorescent microscope and analyzed with the software associated with the microscope. Representative insertions were digitally enlarged and shown below in each image. Please click here to view a larger version of this figure.
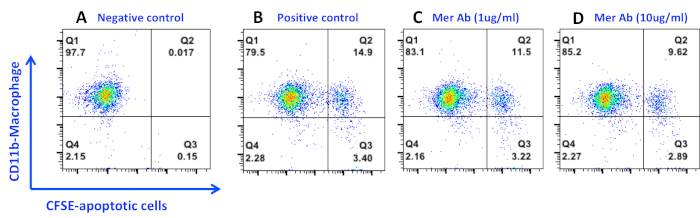
Figure 3. Inhibition of peritoneal macrophage efferocytosis by an anti-Mer antibody. Apoptotic cells were prepared and co-cultured with macrophages for 4 h as described in the protocol. Anti-Mer Ab was added into the macrophage culture immediately before the co-culture with apoptotic cells. Phagocytic macrophages were then detached and stained with anti-mouse CD11b-PE antibody on ice for 20 min. Data were acquired and analyzed as in Figure 1. Please click here to view a larger version of this figure.
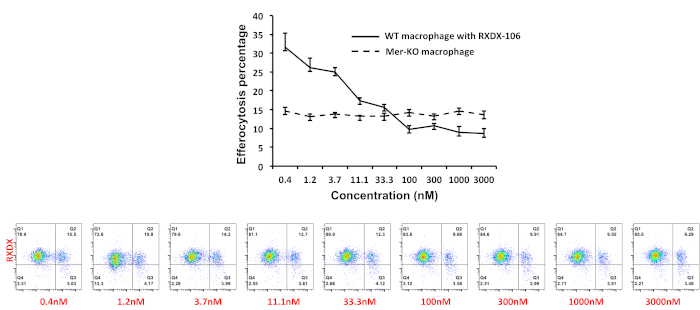
Figure 4. RXDX-106 mediated inhibition of macrophage efferocytosis. Efferocytosis was set up as described in Figure 3. RXDX-106 was added two hrs before the co-culture with apoptotic thymocytes. Mer-deficient peritoneal macrophages were prepared similarly and served as the control group. Data were acquired and the percentage of phagocytic macrophages were gated on CD11b positive cells and analyzed using the flow cytometer software. Please click here to view a larger version of this figure.
Discussion
Apoptosis is a highly conserved cell death process that involves many signal cascades and induces protein expression, secretion, and transportation. Apoptosis is often associated with cellular morphology changes17. Apoptotic cells actively release cytokines and chemokines that attract phagocytes to migrate to the site and initiate the process of engulfment, an extremely complex pathway under tight control18. On the other hand, necrotic cell death releases danger signals that trigger inflammatory responses1. Defective or prolonged clearance of apoptotic cells will lead to secondary necrosis of these cells19. Therefore, timing in apoptosis induction is very critical in the experiment. There are numerous ways to induce cell apoptosis20. However, the duration and strength of induction are cell type dependent. We recommend setting up a titration experiment to determine the optimal condition of maximum (90-95%) apoptosis production. The Annexin-V/7-AAD Apoptotic Detection Kit was used and the instructions were followed in our lab to evaluate the apoptotic efficiency. Our laboratory has tried two different ways to induce thymocytes apoptosis in the past. Gamma-radiation seems to be a better method, as it has no chemical residuals in the culture and induces few necrotic cell deaths. We expose thymocytes to 500 rad of g-radiation followed by 4-h culture in RPMI1640 medium. We observed about 95% apoptotic thymocytes13. When a radiator is not available, we induced thymocytes to undergo apoptosis with 1 μM of staurosporine in the culture for 4 h (Step 1.15). Primary cells are generally more sensitive to apoptosis induction. Extensive washing steps are also required to remove the chemicals from culture before co-culture with phagocytes. There are many ways to label apoptotic cells. Though toxicity has been associated with high concentration, CFSE is very efficiently retained within the cytoplasm when carefully optimized21. pHrodo is an acid-sensitive dye, which increases in fluorescence as the pH of the environment decreases. Due to the low pH of the phagolysosome, phagocytized apoptotic cells can be easily distinguished from physically attached but not engulfed apoptotic cells in the assay22.
Numerous surface receptors (TAM receptors, mannose receptor, integrins (CD11b/CD18), scavenger receptors, Fc receptors, et al) allow phagocytes to distinguish self from pathogens and discriminate the subsequent responses23. Different receptors work together to finish a rather complex process. Blocking receptor(s) likely results in a partial inhibition of phagocytosis. Primary macrophages can be prepared from different resources with various methods (bone marrow derived macrophages, peritoneal macrophages, spleen macrophages, et al.). The advantage of thioglycollate-induced macrophages is the skewed phagocytic phenotype of the macrophages; the disadvantage is that thioglycollate needs to be aged in the dark for at least 3 months. However, thioglycollate solution has a shelf life of 2 years. A constant stock of the solution should overcome this disadvantage. In the preparation of peritoneal macrophages, the trace amount of red blood cells in the peritoneal macrophage collection should not affect the experiment, since they are going to be washed away along with the floating cells after 2 h of macrophage culture. A large amount of red blood cells (visible pellet) may require treatment with ACK lysing buffer. Macrophages tend to adhere tightly to the culture surface. Different methods have been cited in the literature to detach adhesion cells from the surface24. Enzyme-based detachment yields the highest levels of cell recovery but may damage surface receptors, impairing cell function and associated analyses. Alteration of surface receptors may also affect receptor-based flow cytometry analysis. Lidocaine causes cell morphology to change to a more spherical conformation due to the blockade of calcium ion channels25. It is the best way to detach macrophages from the surface with no noticeable damage in our experiment.
Macrophages containing apoptotic cells can be analyzed by flow cytometry-based or microscope-based assays. FACS can be applied to analyze a large number of cells in a short time and can further identify cell subtypes by differential staining with antibodies against the specific surface or cytoplasmic proteins. An optimal concentration (ratio) of apoptotic cell numbers in the co-culture system can also be decided by the FACS analysis. Microscopic analysis has limitations regarding cell numbers and differential staining as compared to the FACS analysis. However, fluorescent microscopy provides in-depth information. Microscopic analysis of macrophage engulfment of apoptotic cells is essential to investigate the capacity and the kinetics of macrophages to ingest apoptotic cells. A time-lapse of the whole ingestion progression may provide detailed information regarding how long it takes to engulf one apoptotic cell, and how many apoptotic cells can a single macrophage ingest simultaneously. Phagocytic macrophages can also be analyzed by Western blot to investigate signaling molecules regulated by the engulfment process. Gene expression profiles associated with this process can be evaluated by real time PCR.
Finally, this protocol provides a basic platform in the experimental analysis of efferocytosis. Other cell types (dendritic cells, mesangial cells, epithelial cells) can also function to uptake apoptotic cells at their residential sites. Those cells have preferences in utilizing different receptors to recognize and initiate the process of apoptotic cell clearance2. The overall phagocytic efficiency may be influenced by several factors, including experimental and cell type specific factors. Variable results reported in the literature are probably due to 1) duration of co-culture; 2) resource and preparation of phagocytes and apoptotic cells; 3) methods to dissociate the apoptotic cells from phagocytes. The protocol described here may apply to the phagocytic potential of other cells. Optimization may be required to maximize the phagocytic potential of target cells. We have analyzed renal mesangial cell phagocytosis of apoptotic thymocytes generated in the same way as described in this protocol. Neutrophils play a key role in the innate immune system through elimination of pathogens. Neutrophil phagocytosis of labeled bacteria or particles can be analyzed by flow cytometry26. However, neutrophils have a very short half-life (6-8 h) and neutrophil phagocytosis of bacteria or fungi occurs within seconds to minutes27,28.
Divulgaciones
The authors have nothing to disclose.
Acknowledgements
Research in the Shao Lab is supported by Research Innovative Award from the College of Medicine and the Junior Faculty Pilot Award from the Department of Internal Medicine, University of Cincinnati and grant DK K01_095067 from NIDDK/NIH.
Materials
| Ack lysing buffer | GIBCO | A10492 | |
| Annexin V/7-AAD | BD Pharmingen | 559763 | |
| Anti-Mer antibody | R&D Systems | BAF591 | |
| CD11b-PE (clone M1/70) | BD Pharmingen | 553311 | |
| CFSE | Invitrogen | C1157 | |
| DMSO | Sigma-Aldrich | D-2650 | |
| EDTA (0.5 mM) | GIBCO | 15575-020 | |
| FACS tubes | BD Biosciences | 352017 | |
| Frosted slides | Fisher Scientific | 12-552-343 | |
| Horse Serum (Heat-inactivated) | Invitrogen | 26050088 | |
| Lidocaine | Sigma-Aldrich | L-5647 | Prepare 1% buffer in 1x PBS |
| PBS, 1x | Corning | 21040CV | |
| RPMI-1640 | Corning | 10040CV | |
| RXDX-106 | Selleck Chemicals | CEP-40783 | |
| Staurosprine (100mg) | Fisher Scientific | BP2541-100 | Add 214.3 ml of DMSO into 100mg to make 1mM stocking solution |
| Thioglycolate Medium Brewer Modified | BD Biosciences | 243010 | Prepare 3% thioglycolate buffer in 1´PBS, autoclaved, and store in the dark for 3 months. |
Referencias
- Shao, W. H., Cohen, P. L. Disturbances of apoptotic cell clearance in systemic lupus erythematosus. Arthritis Research and Therapy. 13 (1), 202 (2011).
- Cohen, P. L. Apoptotic cell death and lupus. Springer Seminars in Immunopathology. 28 (2), 145-152 (2006).
- Wong, R. S. Apoptosis in cancer: from pathogenesis to treatment. Journal of Experimental and Clinical Cancer Research. 30, 87 (2011).
- Baig, S., et al. Potential of apoptotic pathway-targeted cancer therapeutic research: Where do we stand. Cell Death and Disease. 7, 2058 (2016).
- Poon, I. K., Lucas, C. D., Rossi, A. G., Ravichandran, K. S. Apoptotic cell clearance: basic biology and therapeutic potential. Nature Reviews Immunology. 14 (3), 166-180 (2014).
- Qian, Y., Wang, H., Clarke, S. H. Impaired clearance of apoptotic cells induces the activation of autoreactive anti-Sm marginal zone and B-1 B cells. Journal of Immunology. 172 (1), 625-635 (2004).
- Hawkins, L. A., Devitt, A. Current understanding of the mechanisms for clearance of apoptotic cells-a fine balance. Journal of Cell Death. 6, 57-68 (2013).
- Lemke, G. Biology of the TAM receptors. Cold Spring Harbor Perspective in Biology. 5 (11), 009076 (2013).
- Stitt, T. N., et al. The anticoagulation factor protein S and its relative, Gas6, are ligands for the Tyro 3/Axl family of receptor tyrosine kinases. Cell. 80 (4), 661-670 (1995).
- Linger, R. M., Keating, A. K., Earp, H. S., Graham, D. K. TAM receptor tyrosine kinases: biologic functions, signaling, and potential therapeutic targeting in human cancer. Advances in Cancer Research. 100, 35-83 (2008).
- van der Meer, J. H., van der Poll, T., van ‘T Veer, C. TAM receptors, Gas6, and protein S: roles in inflammation and hemostasis. Blood. 123 (16), 2460-2469 (2014).
- Lemke, G., Burstyn-Cohen, T. TAM receptors and the clearance of apoptotic cells. Annal of the New York Academy of Sciences. 1209, 23-29 (2010).
- Shao, W. H., Zhen, Y., Eisenberg, R. A., Cohen, P. L. The Mer receptor tyrosine kinase is expressed on discrete macrophage subpopulations and mainly uses Gas6 as its ligand for uptake of apoptotic cells. Clinical Immunology. 133 (1), 138-144 (2009).
- Scott, R. S., et al. Phagocytosis and clearance of apoptotic cells is mediated by MER. Nature. 411 (6834), 207-211 (2001).
- Klarquist, J., Janssen, E. M. The bm12 Inducible Model of Systemic Lupus Erythematosus (SLE) in C57BL/6 Mice. Journal of Visualized Experiment. (105), e53319 (2015).
- Malawista, A., Wang, X., Trentalange, M., Allore, H. G., Montgomery, R. R. Coordinated expression of tyro3, axl, and mer receptors in macrophage ontogeny. Macrophage (Houst). 3, (2016).
- Elmore, S. Apoptosis: a review of programmed cell death. Toxicologic Pathology. 35 (4), 495-516 (2007).
- Ravichandran, K. S. Find-me and eat-me signals in apoptotic cell clearance: progress and conundrums. Journal of Experimental Medicine. 207 (9), 1807-1817 (2010).
- Rock, K. L., Kono, H. The inflammatory response to cell death. Annual Review in Pathology. 3, 99-126 (2008).
- Roberts, K. M., Rosen, A., Casciola-Rosen, L. A. Methods for inducing apoptosis. Methods in Molecular Medicine. 102, 115-128 (2004).
- Progatzky, F., Dallman, M. J., Lo Celso, C. From seeing to believing: labelling strategies for in vivo cell-tracking experiments. Interface Focus. 3 (3), 20130001 (2013).
- Stijlemans, B., et al. Development of a pHrodo-based assay for the assessment of in vitro and in vivo erythrophagocytosis during experimental trypanosomosis. PLoS Neglected Tropical Diseases. 9 (3), 0003561 (2015).
- Hochreiter-Hufford, A., Ravichandran, K. S. Clearing the dead: apoptotic cell sensing, recognition, engulfment, and digestion. Cold Spring Harbor Perspective in Biology. 5 (1), 008748 (2013).
- Chen, S., So, E. C., Strome, S. E., Zhang, X. Impact of Detachment Methods on M2 Macrophage Phenotype and Function. Journal of Immunology Methods. 426, 56-61 (2015).
- Fleit, S. A., Fleit, H. B., Zolla-Pazner, S. Culture and recovery of macrophages and cell lines from tissue culture-treated and -untreated plastic dishes. Journal of Immunology Methods. 68 (1-2), 119-129 (1984).
- Fine, N., Barzilay, O., Glogauer, M. Analysis of Human and Mouse Neutrophil Phagocytosis by Flow Cytometry. Methods Molecular Biology. 1519, 17-24 (2017).
- Summers, C., et al. Neutrophil kinetics in health and disease. Trends in Immunology. 31 (8), 318-324 (2010).
- Dale, D. C., Boxer, L., Liles, W. C. The phagocytes: neutrophils and monocytes. Blood. 112 (4), 935-945 (2008).

