A Tandem Liquid Chromatography–Mass Spectrometry-based Approach for Metabolite Analysis of Staphylococcus aureus
Summary
Here we describe a protocol for the extraction of metabolites from Staphylococcus aureus and their subsequent analysis via liquid chromatography and mass spectrometry.
Abstract
In an effort to thwart bacterial pathogens, hosts often limit the availability of nutrients at the site of infection. This limitation can alter the abundances of key metabolites to which regulatory factors respond, adjusting cellular metabolism. In recent years, a number of proteins and RNA have emerged as important regulators of virulence gene expression. For example, the CodY protein responds to levels of branched-chain amino acids and GTP and is widely conserved in low G+C Gram-positive bacteria. As a global regulator in Staphylococcus aureus, CodY controls the expression of dozens of virulence and metabolic genes. We hypothesize that S. aureus uses CodY, in part, to alter its metabolic state in an effort to adapt to nutrient-limiting conditions potentially encountered in the host environment. This manuscript describes a method for extracting and analyzing metabolites from S. aureus using liquid chromatography coupled with mass spectrometry, a protocol that was developed to test this hypothesis. The method also highlights best practices that will ensure rigor and reproducibility, such as maintaining biological steady state and constant aeration without the use of continuous chemostat cultures. Relative to the USA200 methicillin-susceptible S. aureus isolate UAMS-1 parental strain, the isogenic codY mutant exhibited significant increases in amino acids derived from aspartate (e.g., threonine and isoleucine) and decreases in their precursors (e.g., aspartate and O-acetylhomoserine). These findings correlate well with transcriptional data obtained with RNA-seq analysis: genes in these pathways were up-regulated between 10- and 800-fold in the codY null mutant. Coupling global analyses of the transcriptome and the metabolome can reveal how bacteria alter their metabolism when faced with environmental or nutritional stress, providing potential insight into the physiological changes associated with nutrient depletion experienced during infection. Such discoveries may pave the way for the development of novel anti-infectives and therapeutics.
Introduction
Bacterial pathogens must contend with many challenges within the host environment. In addition to direct attack by immune cells, the host also sequesters nutrients essential for bacterial survival and replication, generating nutritional immunity1,2. To survive these hostile environments, bacterial pathogens deploy virulence factors. Some of these factors allow the bacteria to evade the immune response; other factors include secreted digestive enzymes, such as hyaluronidase, thermonuclease, and lipase, which may enable the bacteria to replenish missing nutrients by consuming tissue-derived constituents3,4,5. Indeed, bacteria have evolved regulatory systems that tie the physiological state of the cell to the production of virulence factors6,7,8,9,10.
A growing body of evidence points to CodY as a critical regulator linking metabolism and virulence. Although first discovered in Bacillus subtilis as a repressor of the dipeptide permease (dpp) gene11, CodY is now known to be produced by nearly all the low G+C Gram-positive bacteria12,13 and regulates dozens of genes involved in carbon and nitrogen metabolism14,15,16,17,18,19. In pathogenic species, CodY also controls the expression of some of the most important virulence genes20,21,22,23,24,25,26,27. CodY is activated as a DNA-binding protein by two classes of ligands: branched-chain amino acids (BCAAs; isoleucine, leucine, and valine [ILV]) and GTP. When these nutrients are abundant, CodY represses (or in some cases, stimulates) transcription. As these nutrients become limited, CodY activity is progressively reduced, resulting in a graded transcriptional response that re-routes precursors through various metabolic pathways connected to central metabolism28,29,30.
Tandem liquid chromatography coupled to mass spectrometry (LC-MS) is a powerful technique that can accurately identify and quantify small-molecule intracellular metabolites31. When paired with transcriptome analysis (e.g., RNA-Seq), this analytical workflow can provide insight into the physiological changes that occur in response to environmental or nutritional stress. Here, we present a method for metabolite extraction from Staphylococcus aureus cells and subsequent analysis via LC-MS. This approach has been used to demonstrate the pleiotropic effects of CodY on S. aureus physiology.
Protocol
1. Preparation of Buffer Solutions
- Prepare phosphate-buffered saline (PBS; pH 7.4) by diluting a stock solution of 10x PBS to a final concentration of 1x with ultrapure (distilled and deionized) water.
- Prepare quenching solution by combining 2 mL of acetonitrile, 2 mL of methanol, 1 mL of ultrapure H2O, and 19 µL (0.1 mM final concentration) of formic acid.
- Prepare LC-MS solvent A by adding formic acid (0.2% [v/v] final concentration) to ultrapure water.
- Prepare LC-MS solvent B by adding formic acid (0.2% [v/v] final concentration) to acetonitrile.
NOTE: All solutions should be prepared using the highest-purity reagents available (generally high-performance liquid chromatography grade). Solutions should be prepared fresh before each experiment and stored on ice prior to use.
2. Establishment of Steady-state S. aureus Growth
- Streak S. aureus strains of interest for isolation on tryptic soy agar (TSA) from a frozen glycerol stock. Incubate at 37 °C for 16-24 h.
- Inoculate 4 mL of tryptic soy broth (TSB) or another suitable medium in sterile glass incubation tubes with single colonies of each strain. Incubate inclined (~70° angle) with rotation at 60 rotations per min (rpm) at 37 °C for 16-20 h.
NOTE: Overnight cultures are prone to oxygen gradients when using standard methods, including those described in step 2.2, which affect cellular physiology. Thus, we employ a multiple back-dilution strategy to ensure biological steady state (see steps 2.4-3.2, below). - Use a spectrophotometer to measure the optical density of the cultures from step 2.2 at 600 nm (OD600). Use sterile medium as an optical reference (blank). Dilute these cells to an OD600 of 0.05 in 50 mL of sterile TSB medium (pre-warmed to 37 °C) in separate, 250 mL DeLong flasks.
- Incubate the cultures at 37 °C in a water bath with shaking at 280 rpm.
- Every 30 min, take OD600 measurements; as the optical densities increase, it may become necessary to dilute the cultures with TSB so that they remain within the linear absorbance range of the spectrophotometer.
- When cultures from step 2.5 achieve an OD600 of ~0.8-1.0, subculture them into 50 mL of 37 °C TSB to an OD600 of 0.01-0.05 and repeat steps 2.4 and 2.5.
3. Sample Collection Setup
- Prepare a bed of crushed dry ice in an appropriate vessel (e.g., glass dish, ice bucket, or cooler).
- As the optical densities of the cultures approach the desired harvest point, add 1 mL of quenching solution to a 35 mm untreated Petri dish and pre-cool on dry ice for ≥5 min.
NOTE: The "desired harvest point" will vary depending on experimental goals. For example, if one were to examine metabolites during aerobic growth, key indicators of this state include acetate excretion and re-assimilation of the acetate during the post-exponential growth phase32,33. Generally, this point should be within a specific growth stage (e.g., exponential phase). The specific OD600 values associated with this stage may vary between different bacterial strains and growth media. - Place a stainless steel filter frit (pre-chilled to -20 °C) in a rubber stopper and place it atop a vacuum flask attached to a house vacuum or vacuum pump.
- Apply the vacuum and place a mixed cellulose ester membrane (0.22 µm pore size) on top.
NOTE: It is critical to use a filter with a diameter equal to that of the frit and to properly center this filter to ensure that the sample is drawn through the filter rather than over the edge. Wetting the membrane with ice-cold, sterile H2O may help with positioning the membrane.
4. Sample Harvest
- At an OD600 of ~0.4-0.5, use a serological pipette to remove 13 mL of culture from the flask and to apply the sample to the filter.
- After the entire sample has been filtered, immediately wash the filter with ≥5 mL of ice-cold PBS to wash away medium-associated metabolites.
- Disconnect the vacuum and use a pair of sterile tweezers to remove the filter from the frit. Invert the filter (cell-side down) into the pre-chilled quench solution.
NOTE: It is important to perform the above steps quickly (i.e., within seconds) and as soon as the liquid has been removed to ensure the rapid quenching of the cells, arresting metabolic activity. - Incubate the filter in quench solution on dry ice for ≥20 min.
- Using sterile tweezers, invert the filter (cell-side up) in the petri dish and use a micropipette to rinse the cells off of the membrane into the quench solution.
- Re-suspended the cells in quench solution and then transfer the cell suspension to a sterile 2 mL impact-resistant tube containing ~100 µL of 0.1 mm silica beads. Store this on dry ice or at -80 °C.
5. Metabolite Extraction
- Thaw samples on wet ice and disrupt the cells in a homogenizer with four 30 s bursts at 6,000 rpm, with 2 min cooling periods on dry ice between cycles.
- Clarify the lysates for 15 min in a pre-chilled, refrigerated microcentrifuge at maximum speed (i.e., 18,213 x g at ≤4 °C).
- Transfer the supernatant to a clean microcentrifuge tube.
- Using a micropipette, transfer a small portion of the sample to a microcentrifuge tube for the quantification of residual peptide content in step 6; store the remainder at -80 °C.
NOTE: The volume of reserved sample varies, depending on the BCA assay used in step 6.1. This sample should be stored on wet ice for immediate analysis or frozen at -80 °C.
6. Bicinchoninic Acid (BCA) Assay
- Perform a BCA assay as recommended by the kit manufacturer, using samples from step 5.4 to determine the residual peptide concentration for each sample.
7. LC-MS
- Mix 75 µL of S. aureus extract with 75 µL of LC-MS solvent B, prepared in step 1.4.
- Vortex to mix and spin at 13,000 x g for 5 min.
- Place 100 µL of supernatant into a liquid chromatography (LC) vial and cap it. Ensure that no air bubbles are trapped in the sample.
- Load the LC vials onto the LC-MS autosampler and edit the running list in the software "Offline Worklist Editor."
- Fill out the "Sample Name" (e.g., wild-type-1), "Sample Position" (e.g., P1-A1), "Method" (e.g., Formic Acid-Negative Method), and "Data File" (e.g., wild-type-1) columns. Click the button "Save Worklist" button. Open the "Mass Spectrometry Data Acquisition Workstation" software and input the previously saved worklist. Click the "Start Worklist Run" button to start the continuous LC-MS measurement.
- Separate the samples on a column, link the column to a time of flight (TOF) spectrometer, and couple the TOF spectrometer with the LC system. Use a mobile-phase gradient as follows: 0-2 min, 85% solvent B; 3-5 min, 80% solvent B; 6-7 min, 75% solvent B; 8-9 min, 70% solvent B; 10-11.1 min, 50% solvent B; 11.1-14 min, 20% solvent B; and 14.1-24 min, 5% solvent B; end with a 10 min re-equilibration period at 85% solvent B and a flow rate of 0.4 mL min-1.
- Using an isocratic pump, infuse a reference mass solution with the run to allow for simultaneous mass axis calibration.
NOTE: This step is based on the standard TOF spectrometer manual.- Use the mixture of acetic acid D4 and hexakis (1H,1H,3H-tetrafluoropropoxy) phosphazine as the reference mass solution to perform the real-time calibration. Use the isocratic pump with the flow rate of 2.5 mL min-1 for the infusion.
8. Batch Correction of Ion Counts
- Designate any sample to serve as a reference sample for batch correction (e.g., wild-type, replicate 1).
- Calculate the sum of the ion counts for all metabolites within the reference sample. Repeat this calculation for all samples.
- Divide the total ion count of each sample by the total ion count of the reference sample to generate a ratio.
- Divide the ion count for each metabolite within a sample by the sample/reference ratio to obtain a batch-corrected ion count for each metabolite.
9. Peptide Normalization
- Divide the batch-corrected ion count values for each sample obtained in step 8 by the peptide concentration determined with the BCA assay in step 6 to yield a normalized value for each metabolite.
NOTE: The normalized, batch-batch corrected ion counts for each metabolite obtained in step 9.1 can be directly compared between strains and subjected to statistical analysis (e.g., a Mann-Whitney U-test). Alternatively, a metabolite known to be unchanged either by the treatment or genetic background may be used as a normalizer to detect changes due to metabolite decomposition. The inclusion of a known amount of L-norvaline or gluraric acid in the extraction buffer can be used to correct for loss during sample processing34.
Representative Results
We have analyzed intracellular metabolite pools in S. aureus during in vitro growth in a rich, complex medium. As proof of principle, we compared metabolite profiles between the methicillin-susceptible S. aureus osteomyelitis isolate UAMS-1 (wild-type [WT]) and an isogenic strain lacking the global transcriptional regulator CodY (ΔcodY)26. Steady-state, exponential cultures of the WT and codY strains were established in TSB medium, as described in step 2 of the protocol. The growth behavior of the wild-type and codY-null mutant cultures were similar, with only mild differences in growth yield and rate (Figure 1). Using RNA-Seq and microarray technology, we and others revealed that multiple genes coding for enzymes involved in the biosynthesis of amino acids derived from aspartate were de-repressed in the codY-null mutant compared to WT cells during in vitro growth in TSB (Figure 2)25,27,30. Moreover, brnQ1 and brnQ2, which code for branched-chain amino acid permeases, are overexpressed in the codY-null mutant30,35.
To determine the extent to which steady-state intracellular abundances of metabolites associated with this pathway are altered in the null mutant, we performed LC-MS-based metabolite profiling. WT and codY-null mutant cells were grown to biological steady state and were sampled as described in step 4 of the protocol. We determined metabolite abundances by integrating the peak area ion intensity for each chromatographically resolved metabolite using an analytical software package (see Materials List). We corrected for differences in biomass by normalizing metabolite abundances to the residual peptide concentration of each sample. We further corrected these values for potential batch effects between samples by calculating the average ion count for all metabolites within each sample and by using the wild-type sample as the reference value. This approach enabled inter-sample comparisons of metabolite abundances across conditions. Inter-metabolite comparisons within a given sample can similarly be achieved by first converting normalized metabolite abundances from ion counts to molar quantities using the method of standard addition.
We have compared the levels of key intermediates in the aspartate pathway in UAMS-1 and its codY-null mutant. As seen in Figure 3, the end products of this pathway (e.g., threonine and (iso)-leucine) are more abundant in codY-null mutant cells, while precursors (e.g., aspartate and o-acetyl homoserine) are more abundant in WT cells. The combined upregulation of BrnQ permeases36 and the ILV biosynthetic pathway likely leads to increases in isoleucine and leucine30. Although the differences are relatively small (<4-fold), LC-MS-based quantitation and batch correction reveal robust and statistically significant changes that are consistent with transcriptional alterations mediated by CodY.
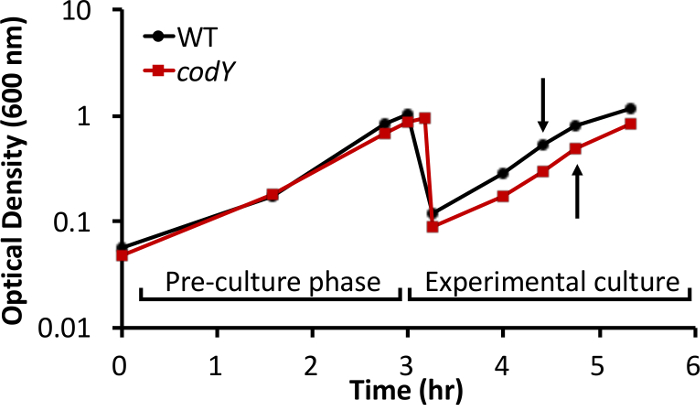
Figure 1: Growth behavior of S. aureus UAMS-1 and an isogenic codY-null mutant in TSB. To extend the time the cells spent in steady-state exponential growth, cultures were back-diluted to an optical density of 0.05 in fresh medium after the pre-cultures achieved an OD600 of ~1. Samples for LC-MS metabolite analysis were collected from experimental cultures at an optical density of ~0.5 (arrows). The data shown are representative of three biological replicates.
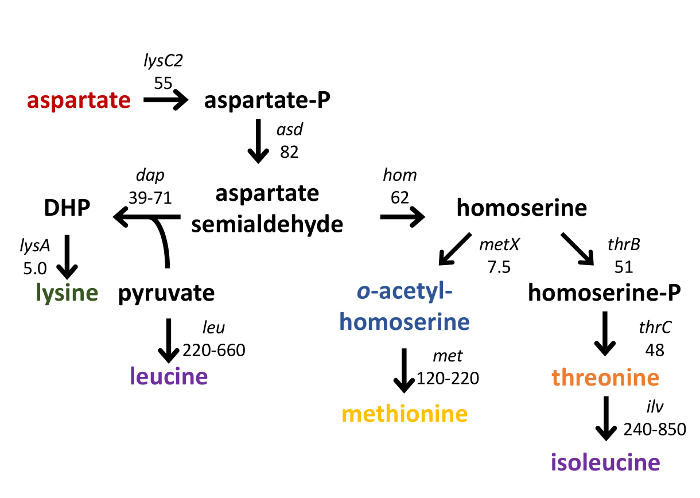
Figure 2: Schematic of selected metabolites derived from aspartate. Genes and operons whose products catalyze the synthesis of aspartate-family amino acids are indicated in italics; the increase in transcript abundance in a codY-null mutant compared to wild type, as determined by RNA-seq analysis29, is also noted. DHP, 2,6-diaminoheptanedioate (2,6-diaminopimelate).
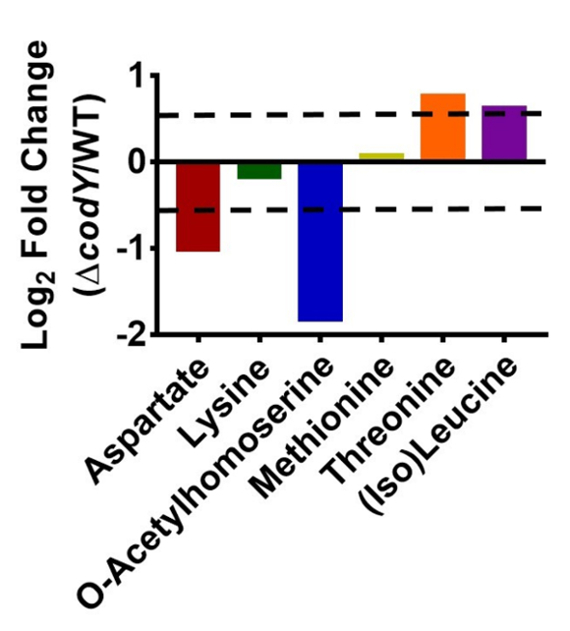
Figure 3: Abundances of metabolites in the aspartate family are altered in a codY mutant. The log2-fold changes of selected metabolites in the codY-null mutant compared to UAMS-1 (WT) are shown. The change was determined by dividing the average abundance of three biological replicates of the codY-null strain by the average abundance of three biological replicates of the WT strain. The standard error between biological replicates for each metabolite was <35%. The dashed lines indicate a log2 1.5-fold change, the cutoff used in this experiment.
Discussion
All small-molecule metabolites are connected to one another through their common origins in central metabolic pathways. During exponential growth, bacterial cells are at biological and metabolic steady state, providing a snapshot of the physiological state under specific conditions. CodY monitors nutrient sufficiency by responding to ILV and GTP. As ILV and GTP pools drop, CodY activity is likely progressively reduced, adjusting the expression of its target genes to adapt to increasing nutrient depletion30. A CodY-deficient strain behaves as though ILV and GTP are depleted from the environment but only exhibits a very mild difference in growth behavior relative to the CodY-proficient strain (Figure 1). Thus, the comparison of steady-state pools of metabolites in these strains provides us with a unique opportunity to reveal the extent to which metabolism is reconfigured when nutrients are scarce. It should be noted that our experiments investigate metabolite abundances when CodY activity is maximized (ILV and GTP are most abundant in exponential phase). However, other questions may be addressed in other phases of growth. For example, in exponential phase, tricarboxylic acid (TCA) cycle activity is very low; in post-exponential phase, the TCA cycle is activated36. A number of phenotypes, including metabolite abundances, are dependent upon this activation. Additionally, the agr quorum sensing system becomes active during the transition from exponential growth to stationary phase37. Collecting samples in post-exponential or stationary phase may be more relevant for studies addressing these topics. Regardless of when the samples are harvested, it is important to collect, wash, and transfer the samples to the extraction buffer as quickly as possible (i.e., within s) to minimize the metabolite turnover that occurs when steady state is perturbed.
The chromatographic method described here is especially suited to analyzing polar and non-polar amino acids and central carbon metabolism compounds. However, additional class-specific methods can be used to quantify specific compounds of interest not chromatographically resolved by this method. For example, altering the pH of the chromatographic mobile phase by replacing formic acid with acetic acid as an additive has been reported to enable the resolution of isoleucine and leucine38. Modifying the extraction procedure can similarly enable the recovery and quantification of labile metabolites that are sensitive to the extraction method used here. For example, compounds such as cysteine that are prone to disulfide bond formation can potentially be recovered following derivatization with Ellman's reagent39. Sparging extraction buffers with nitrogen gas can preserve NAD+ and NADH ratios, allowing for an assessment of the cellular redox state40. Nucleoside triphosphates can decompose in basic or unbuffered solutions; acidifying the extraction solution can improve the recovery of these molecules41.
In an exponentially growing bacterial culture, the depletion of one or more nutrients leads to the transition to the post-exponential and stationary phases of growth. These growth phases are characterized by distinct metabolic states42. Chemostat-based cultures generate a virtually continuous biological steady state ideal for gene expression and physiological studies. However, the method requires specialized equipment, is technically demanding, and necessitates that a nutrient limitation be imposed to maintain a stable population of cells under flow. The latter requirement causes transcriptional and physiological perturbations due to factors other than the variable or regulator being analyzed. To ensure that our flask-based results are representative of cells growing exponentially at steady state and not of a transition between two phases, we employ a double-back dilution strategy with a consistent flask:volume ratio (slight changes in oxygen levels can lead to altered metabolism36,42). After an initial dilution of overnight cultures, these cells are grown to an OD600 of ~1.0, back-diluted to an OD600 of ~0.05, and harvested when they reach an OD600 of ~0.5. Such a method also dilutes cytoplasmic molecules that accumulate during overnight growth, including stable RNAs. Indeed, RNAIII, the effector of the agr quorum sensing system, is one such RNA and regulates the expression of some of the same gene targets as CodY25,43,44. Accumulated RNAIII can mask CodY-dependent regulation, leading to an underestimate of the strength of repression or stimulation by CodY (Sharma and Brinsmade, unpublished results).
One limitation of this analysis is that it provides an instantaneous glimpse of metabolite abundances within the cell; no conclusions can be drawn from the results regarding changes in flux through any given pathway. For example, the abundances of lysine and methionine between the two strains examined did not change, despite de-repression of biosynthetic enzymes in the codY-null strain (Figures 2 and 3). The codY-null strain may in fact be generating more lysine and methionine, but they may be rapidly converted into other compounds; thus, these molecules do not accumulate. Using 13C- or 15N-labeled carbon or nitrogen sources would allow us to follow carbon and nitrogen skeletons through major metabolic junctions45,46.
We have used the described method to elucidate changes in metabolite pools in S. aureus, B. subtilis29, Mycobacterium tuberculosis47, and Enterococcus faecium48, but the method may be applied to other Gram-positive and Gram-negative bacteria, including other human pathogens easily cultivated in the laboratory. Indeed, integrating metabolomic and transcriptomic information can reveal unexpected connections between metabolism and virulence, which could lead to novel strategies to treat infections.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was funded in part by an NIH Pathway to Independence Award (grant GM 099893) and faculty startup funds to SRB, as well as a Research Project Grant (grant GM 042219). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Materials
| Material/Equipmenta | |||
| DeLong Culture Flask (250 ml) | Belco | 2510-00250 | |
| Sidearm Flask, 500 ml | Pyrex | 5340 | |
| 3-hole Rubber Stopper, #7 | Fisher | 14-131E | |
| Stainless Steel Filter holder/frit | VWR | 89428-936 | |
| Petri Dish, 35 mm | Corning | 430588 | Not tissue culture treated |
| Mixed cellulose ester membrane, 0.22 μm pore size | Millipore | GSWP02500 | |
| Impact-resistant tubes, 2 ml | USA Scientific | 1420-9600 | |
| Silica Beads, 0.1 mm | Biospec Products Inc | 11079101Z | |
| Precellys 24 homogenizer | Bertin Instruments | EQ03119-200-RD000.0 | |
| Micro BCA Protein Assay Kit | Pierce (Thermo Scientific) | 23235 | |
| Cogent Diamond hydride type C column | Agilent | 70000-15P-2 | |
| Accurate-Mass Time-of-Flight (TOF) LC-MS, 6200 Series | Agilent | G6230B | |
| Quat Pump, 1290 Series | Agilent | G4204A | |
| Bin Pump, 1290 Series | Agilent | G4220A | |
| Valve Drive, 1290 Series | Agilent | G1107A | |
| Isocratic Pump, 1290 Series | Agilent | G1310B | |
| TCC, 1290 Series | Agilent | G1316C | |
| Sampler, 1290 Series | Agilent | G4226A | |
| Thermostat, 1290 Series | Agilent | G1330B | |
| Name | Company | Catalog Number | Comments |
| Chemical | |||
| Tryptic Soy Broth | Becton Dickinson | 211825 | |
| Difco Agar, Granulated | Becton Dickinson | 214530 | Solid media contains 1.5% [w/v] agar |
| Phosphate-buffered saline (pH 7.4) 10X | Ambion | AM9624 | Dilute fresh to 1X with ultra-pure water |
| Acetonitrile | Fisher Scientific | A955-500 | Optima LC-MS |
| Methanol | Fisher Scientific | A456-500 | Optima LC-MS; toxic |
| Formic Acid | Sigma Aldrich | 94318 | For mass spectrometry, 98% |
| Name | Company | Catalog Number | Comments |
| Software | |||
| MassHunter | Agilent | G3337AA | |
| Bacterial Strain | Species | Strain | Genotype |
| SRB 337 | Staphylococcus aureus | USA200 MSSA UAMS-1 | wild type |
| SRB 372 | Staphylococcus aureus | USA200 MSSA UAMS-1 | ΔcodY::erm |
| aChemicals and materials listed are specific to the method described and do not include standard laboratory chemicals or supplies. |
References
- Hood, M. I., Skaar, E. P. Nutritional immunity: transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10 (8), 525-537 (2012).
- Weinberg, E. D. Clinical enhancement of nutritional immunity. Comp. Ther. 1 (5), 38-40 (1975).
- Ibberson, C. B., et al. Staphylococcus aureus hyaluronidase is a CodY-regulated virulence factor. Infect. Immun. 82 (10), 4253-4264 (2014).
- Lee, C. Y., Iandolo, J. J. Mechanism of bacteriophage conversion of lipase activity in Staphylococcus aureus. J. Bacteriol. 164 (1), 288-293 (1985).
- Olson, M. E., et al. Staphylococcus aureus nuclease is an SaeRS-dependent virulence factor. Infect. Immun. 81 (4), 1316-1324 (2013).
- Somerville, G. A., Proctor, R. A. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol. Mol. Biol. Rev. 73 (2), 233-248 (2009).
- Seidl, K., et al. Staphylococcus aureus CcpA affects virulence determinant production and antibiotic resistance. Antimicrob. Agents Chemother. 50 (4), 1183-1194 (2006).
- Richardson, A. R., Somerville, G. A., Sonenshein, A. L. Regulating the intersection of metabolism and pathogenesis in Gram-positive bacteria. Microbiol. Spectr. 3 (3), 1-27 (2015).
- Geiger, T., et al. Role of the (p)ppGpp synthase RSH, a RelA/SpoT homolog, in stringent response and virulence of Staphylococcus aureus. Infect. Immun. 78 (5), 1873-1883 (2010).
- Gaupp, R., et al. RpiRc is a pleiotropic effector of virulence determinant synthesis and attenuates pathogenicity in Staphylococcus aureus. Infect. Immun. 84 (7), 2031-2041 (2016).
- Serror, P., Sonenshein, A. L. Interaction of CodY, a novel Bacillus subtilis DNA-binding protein, with the dpp promoter region. Mol. Microbiol. 20 (4), 843-852 (1996).
- Sonenshein, A. L. CodY, a global regulator of stationary phase and virulence in Gram-positive bacteria. Curr. Opin. Microbiol. 8 (2), 203-207 (2005).
- Brinsmade, S. R. CodY, a master integrator of metabolism and virulence in Gram-positive bacteria. Curr. Genet. , (2016).
- Molle, V., et al. Additional targets of the Bacillus subtilis global regulator CodY identified by chromatin immunoprecipitation and genome-wide transcript analysis. J. Bacteriol. 185 (6), 1911-1922 (2003).
- Moses, S., et al. Proline utilization by Bacillus subtilis: Uptake and catabolism. J. Bacteriol. 194 (4), 745-758 (2012).
- Lobel, L., Herskovits, A. A. Systems level analyses reveal multiple regulatory activities of CodY controlling metabolism, motility, and virulence in Listeria monocytogenes. PLoS Genet. 12 (2), 1-27 (2016).
- Belitsky, B. R., Sonenshein, A. L. CodY-mediated regulation of guanosine uptake in Bacillus subtilis. J. Bacteriol. 193 (22), 6276-6287 (2011).
- den Hengst, C. D., Buist, G., Nauta, A., Van Sinderen, D., Kuipers, O. P., Kok, J. Probing direct interactions between CodY and the oppD promoter of Lactococcus lactis. Microbiol. 187 (2), 512-521 (2005).
- Fisher, S. H. Regulation of nitrogen metabolism in Bacillus subtilis: vive la différence. Mol. Microbiol. 32 (2), 223-232 (1999).
- Dineen, S. S., McBride, S. M., Sonenshein, A. L. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 192 (20), 5350-5362 (2010).
- Dineen, S. S., Villapakkam, A. C., Nordman, J. T., Sonenshein, A. L. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66 (1), 206-219 (2007).
- Hendriksen, W. T., et al. CodY of Streptococcus pneumoniae: Link between nutritional gene regulation and colonization. J. Bacteriol. 190 (2), 590-601 (2008).
- Bennett, H. J., et al. Characterization of relA and codY mutants of Listeria monocytogenes: Identification of the CodY regulon and its role in virulence. Mol. Microbiol. 63 (5), 1453-1467 (2007).
- Stenz, L., Francois, P., Whiteson, K., Wolz, C., Linder, P., Schrenzel, J. The CodY pleiotropic repressor controls virulence in Gram-positive pathogens. FEMS Immunol. and Med. Microbiol. 62 (2), 123-139 (2011).
- Majerczyk, C. D., et al. Direct targets of CodY in Staphylococcus aureus. J. Bacteriol. 192 (11), 2861-2877 (2010).
- Majerczyk, C. D., Sadykov, M. R., Luong, T. T., Lee, C., Somerville, G. A., Sonenshein, A. L. Staphylococcus aureus CodY negatively regulates virulence gene expression. J. Bacteriol. 190 (7), 2257-2265 (2008).
- Pohl, K., et al. CodY in Staphylococcus aureus: A regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191 (9), 2953-2963 (2009).
- Sonenshein, A. L. Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5 (12), 917-927 (2007).
- Brinsmade, S. R., et al. Hierarchical expression of genes controlled by the Bacillus subtilis global regulatory protein CodY. Proc. Nat. Acad. Sci. U.S.A. 111 (22), 2-7 (2014).
- Waters, N. R., et al. A spectrum of CodY activities drives metabolic reorganization and virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 101 (3), 495-514 (2016).
- Zhou, B., Xiao, J. F., Tuli, L., Ressom, H. W. LC-MS-based metabolomics. Mol. Biosyst. 8 (2), 470-481 (2012).
- Somerville, G. A., et al. Staphylococcus aureus aconitase inactivation unexpectedly inhibits post-exponential-phase growth and enhances stationary-phase survival. Infect. Immun. 70 (11), 6373-6382 (2002).
- Somerville, G. A., Said-Salim, B., Wickman, J. M., Raffel, S. J., Kreiswirth, B. N., Musser, J. M. Correlation of acetate catabolism and growth yield in Staphylococcus aureus: Implications for host-pathogen interactions. Infect. Immun. 71 (8), 4724-4732 (2003).
- Brinsmade, S. R., Kleijn, R. J., Sauer, U., Sonenshein, A. L. Regulation of CodY activity through modulation of intracellular branched-chain amino acid pools. J. Bacteriol. 192 (24), 6357-6368 (2010).
- Kaiser, J. C., Omer, S., Sheldon, J. R., Welch, I., Heinrichs, D. E. Role of BrnQ1 and BrnQ2 in branched-chain amino acid transport and virulence in Staphylococcus aureus. Infect. Immun. 83 (3), 1019-1029 (2015).
- Ledala, N., Zhang, B., Seravalli, J., Powers, R., Somerville, G. A. Influence of iron and aeration on Staphylococcus aureus growth, metabolism, and transcription. J. Bacteriol. 196 (12), 2178-2189 (2014).
- Novick, R. P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Micorbiol. 48 (6), 1429-1449 (2003).
- Pesek, J. J., Matyska, M. T., Fischer, S. M., Sana, T. R. Analysis of hydrophilic metabolites by high-performance liquid chromatography-mass spectrometry using a silica hydride-based stationary phase. J. Chromatog. A. 1204 (1), 48-55 (2008).
- Guan, X., Hoffman, B., Dwivedi, C., Matthees, D. P. A simultaneous liquid chromatography/mass spectrometric assay of glutathione, cysteine, homocysteine and their disulfides in biological samples. J. Pharm. Biomed. Anal. 31 (2), 251-261 (2003).
- Sporty, J. L., Kabir, M. M., Turteltaub, K. W., Ognibene, T., Lin, S. J., Bench, G. Single sample extraction protocol for the quantification of NAD and NADH redox states in Saccharomyces cerevisiae. J. Sep. Sci. 31 (18), 3202-3211 (2008).
- Rabinowitz, J. D., Kimball, E. Acidic acetonitrile for cellular metabolome extraction from Escherichia coli. Anal. Chem. 79 (16), 6167-6173 (2007).
- Somerville, G. A., Powers, R. Growth and preparation of Staphylococcus epidermidis for NMR metabolomic analysis. Methods Mol. Biol. 1106, 71-91 (2014).
- Roux, A., Todd, D. A., Velazquez, J. V., Cech, N. B., Sonenshein, A. L. CodY-Mediated regulation of the Staphylococcus aureus Agr system integrates nutritional and population density signals. J. Bacteriol. 196 (6), 1184-1196 (2014).
- Guillet, J., Hallier, M., Felden, B. Emerging functions for the Staphylococcus aureus RNome. PLoS Pathog. 9 (12), 1003767 (2013).
- Sauer, U., et al. Metabolic flux ratio analysis of genetic and environmental modulations of Escherichia coli central carbon metabolism. J. Bacteriol. 181 (21), 6679-6688 (1999).
- Niittylae, T., Chaudhuri, B., Sauer, U., Frommer, W. B. Comparison of Quantitative Metabolite Imaging Tools and Carbon-13 Techniques for Fluxomics. Methods Mol. Biol. 553 (1), 355-372 (2009).
- de Carvalho, L. P. S., Fischer, S. M., Marrero, J., Nathan, C., Ehrt, S., Rhee, K. Y. Metabolomics of Mycobacterium tuberculosis reveals compartmentalized co-catabolism of carbon substrates. Chem. Biol. 17 (10), 1122-1131 (2010).
- Weisenberg, S. A., Butterfield, T. R., Fischer, S. M., Rhee, K. Y. Suitability of silica hydride stationary phase, aqueous normal phase chromatography for untargeted metabolomic profiling of Enterococcus faecium and Staphylococcus aureus. J. Sep. Sci. 32 (13), 2262-2265 (2009).

