Utilizing a Comprehensive Immunoprecipitation Enrichment System to Identify an Endogenous Post-translational Modification Profile for Target Proteins
Summary
Investigating multiple, endogenous post-translational modifications for a target protein can be extremely challenging. Techniques described here utilize an optimized lysis buffer and filter system developed with specific PTM-targeting, affinity matrices to detect the acetylation, ubiquitination, SUMOylation 2/3, and tyrosine phosphorylation modifications for a target protein using a single, streamlined system.
Abstract
It is now well-appreciated that post-translational modifications (PTMs) play an integral role in regulating a protein's structure and function, which may be essential for a given protein's role both physiologically and pathologically. Enrichment of PTMs is often necessary when investigating the PTM status of a target protein, because PTMs are often transient and relatively low in abundance. Many pitfalls are encountered when enriching for a PTM of a target protein, such as buffer incompatibility, the target protein antibody is not IP-compatible, loss of PTM signal, and others. The degree of difficulty is magnified when investigating multiple PTMs like acetylation, ubiquitination, SUMOylation 2/3, and tyrosine phosphorylation for a given target protein. Studying a combination of these PTMs may be necessary, as crosstalk between PTMs is prevalent and critical for protein regulation. Often, these PTMs are studied in different lysis buffers and with unique inhibitor compositions. To simplify the process, a unique denaturing lysis system was developed that effectively isolates and preserves these four PTMs; thus, enabling investigation of potential crosstalk in a single lysis system. A unique filter system was engineered to remove contaminating genomic DNA from the lysate, which is a problematic by-product of denaturing buffers. Robust affinity matrices targeting each of the four PTMs were developed in concert with the buffer system to maximize the enrichment and detection of the endogenous states of these four PTMs. This comprehensive PTM detection toolset streamlines the process of obtaining critical information about whether a protein is modified by one or more of these PTMs.
Introduction
Post-translational modifications (PTMs) are highly regulated alterations to a protein, whereby the modification is added or removed in a specific manner. PTMs are often dynamic, transient changes that significantly alter the protein's structure, interactions with partner proteins, and spatial localization, and ultimately enabling the protein to perform distinct functions1,2,3,4. PTMs are so abundant that they increase the number of unique protein forms (or proteoforms) from about 30,000 gene products to over a million proteoforms5,6. Identifying PTMs and defining their effect on a target protein is critical towards understanding the protein's physiological and pathological functions7,8,9,10,11,12,13,14. Work on select proteins like tubulin, p53, and epidermal growth factor receptor (EGFR) have elucidated the regulatory roles of PTMs15,16,17. These studies uncovered the fact that regulation of these proteins occurs by multiple PTMs, and in many cases these modifications work in concert to promote a specific function. New studies have shown that both cooperative and negative PTM crosstalk can occur on several different proteins18,19,20,21,22,23,24,25. However, the PTM profiles of the majority of proteins are built from several studies that have used different models and unique conditions. Having an optimized system to investigate multiple PTMs in one system would be highly beneficial to gain insight into potential PTM crosstalk for a target protein.
One challenge with investigating PTMs in a single system is that specific PTMs are investigated using distinct lysis buffers. For example, the utilization of buffers like RIPA or NP-40 that are commonly used for phosphorylation or ubiquitination investigations may be inadequate for studying a labile PTM like small ubiquitin-like modifier (SUMO)ylation26. Additionally, non-denaturing buffers are inadequate for dissociating protein interactions, and can lead to false positive PTM identification27. Investigating PTMs in a denaturing lysis buffer may be preferred, as it is significantly better at isolating proteins from all cellular compartments28, will dissociate most protein interactions, and will inhibit proteases that alter PTM states, such as deSUMOylases 26; however, denaturing buffers may compromise the integrity of specific affinity reagents such as ubiquitin binding domain-based tools27. Developing a denaturing-like lysis system to study multiple PTMs of a protein in an affinity reagent-compatible system would be highly beneficial for PTM crosstalk investigations.
Although denaturing buffers have key advantages over non-denaturing buffers for investigating PTMs, they are less commonly used because of their significant drawbacks, such as incompatibility with conventional protein assays, significant genomic deoxyribonucleic acid (DNA) contamination, and disruption to immunoprecipitation (IP) reagents29. These drawbacks can result in extended preparation time and potential damage to target proteins when shearing genomic DNA. Two commonly used methods to shear DNA include syringe lysis and sonication29. Both methods require extensive optimization and often lack precise reproducibility. Alternative methods to remove genomic DNA include the addition of DNAses; however, this may require additional optimization and changes in buffer composition. Ultimately, a simple and reproducible method to remove genomic DNA contamination would be beneficial when working with denaturing lysis buffers for PTM investigations.
The goal of this technique is to develop a system to isolate and investigate ubiquitination (Ub), tyrosine phosphorylation (pY), SUMOylation 2/3 (SUMO 2/3), and acetylation (Ac) PTMs in a single system to better investigate PTM crosstalk for a target protein. This PTM detection system was utilized to rapidly investigate the endogenous PTM profile of several target proteins in a single system. New insight into potential crosstalk was identified30,31. Overall, the technique described here improves on existing methods to investigate PTMs and PTM crosstalk in three distinct ways: 1) a unique denaturing buffer was developed that isolates proteins from all cellular compartments, while still being compatible with PTM affinity reagents, 2) a unique filter system was developed that rapidly removes genomic DNA contamination from denatured lysates with high reproducibility and requires no specialized equipment, and 3) the robust affinity beads, lysis system, and inhibitory reagents are compatible; thus, simplifying the isolation of these four PTMs for a target protein to maximize PTM enrichment, and efficiently examine potential crosstalk.
Protocol
1. Sample Preparation: Cell Culture
- Grow four 150 mm plates of A431 cells in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum.
- Grow the A431 cells to 50% confluency.
- Serum restrict the four plates of A431 cells with serum-free DMEM for 24 h.
- Treat two plates of A431 cells with 33.3 mg/mL of epidermal growth factor for 1 h. Leave the other two plates untreated.
NOTE: Cell growth and treatment conditions shown here provide an example; however, this methodology is applicable to many different cell types and treatment conditions.
- Prepare blastR lysis and dilution buffers with inhibitors (Table 1). Combine 2.895 mL of blastR lysis buffer with 15 µL of tyrosine phosphatase inhibitor, 30 µL of deubiquitinase and deSUMOylase inhibitor, 30 µL of histone deacetylase (HDAC) inhibitor, and 30 µL of protease inhibitor cocktail.
- Combine 9.65 mL of blastR dilution buffer with 50 µL of tyrosine phosphatase inhibitor, 100 µL of deubiquitinase and deSUMOylase inhibitor, 100 µL of histone deacetylase (HDAC) inhibitor, and 100 µL of protease inhibitor cocktail.
NOTE: See Table of Materials for buffer composition and inhibitor information.
- Combine 9.65 mL of blastR dilution buffer with 50 µL of tyrosine phosphatase inhibitor, 100 µL of deubiquitinase and deSUMOylase inhibitor, 100 µL of histone deacetylase (HDAC) inhibitor, and 100 µL of protease inhibitor cocktail.
- Pour off the culture media and place the cells on ice. Then, wash the cells twice with 10 mL of 1x phosphate buffered saline (PBS).
- Remove as much PBS as possible prior to adding blastR lysis buffer to maximize cell lysis.
NOTE: See Table of Materials for buffer composition.
- Remove as much PBS as possible prior to adding blastR lysis buffer to maximize cell lysis.
- Add 600 µL of blastR lysis buffer for each 150 mm plate. The amount of buffer is based on expected protein yield (Table 2).
- Lyse the cells using a cell scraper. The lysate will become viscous due to nuclear lysis.
- Use a snipped 1 mL pipette tip to transfer the crude lysate into a blastR filter that has been placed in a 15 mL collection tube (Figure 1). Here, a 15 mL falcon tube is used.
- Use a supplied filter plunger to completely compress the blastR filter and collect the lysate flow-through, including any bubbles, in a 15 mL tube (Figure 1).
- Optional: Transfer clarified lysate to a 1.5 mL microcentrifuge tube and centrifuge the lysate at approximately 10,000 x g for 1 min at 4 °C.
- Transfer the supernatant to a new 15 mL falcon tube.
- Dilute the clarified lysate with blastR dilution buffer to a final volume of 3 mL for each 150 mm plate. The final volume is based on expected protein yield (Table 2).
NOTE: This step is important as the final buffer composition will affect the IP reaction stringency. - Quantitate protein concentration (section 2).
2. Protein Quantitation Assay
NOTE: Utilize a standard colorimetric protein quantitation assay to determine protein quantitation. Here, protein quantitation is determined using precision red advanced protein assay.
- Add 1 mL of precision red advanced protein assay reagent to each of two 1 mL cuvettes.
- Mix 10 µL of blastR lysis buffer and 40 µL of blastR dilution buffer in a clean 1.5 mL microcentrifuge tube on ice.
NOTE: This will be used for reading the blank protein sample. - Add 20 µL of the lysis/dilution buffer mix (from step 2.2) to the first cuvette and mix by inverting two to three times.
- Add 20 µL of the diluted cell lysate (from experiment) to the second cuvette, mixing as above.
- Incubate samples for 1 min at room temperature.
- Blank spectrophotometer with the lysis/dilution buffer mix sample (from step 2.3).
- Measure absorbance of the lysate sample (from step 2.4) at 600 nm.
- Determine the lysate protein concentration as follows;
sample reading OD600 x 5 = protein concentration in mg/mL
NOTE: OD readings below 0.05 or above 0.5 are close to the linear range capacity of the protein assay. For readings >0.5, samples can be diluted. For readings <0.05, more lysate can be added to the precision red advanced protein assay reagent (up to 50 µL). See Table 3 for multipliers to convert spectrophotometer readings to mg/mL lysate protein concentration. If there is insufficient protein in one plate, it is recommended to use 2 or more plates per IP. In this case, plates will be harvested in series, transferring the original 300 µL of lysis buffer between plates. - Based on protein concentration, dilute sample with a buffer mix (1 part blastR lysis: 4 parts blastR dilution) to a desired final concentration (usually 1 mg/mL).
- Snap freeze aliquots of samples that will not be used immediately.
- Store samples at -70 °C.
- Proceed to Section 3.
3. Immunoprecipitation (IP) Assay
NOTE: Affinity bead concentrations, lysate concentrations, and incubation times are recommended guidelines, and may be unique for each target protein and specific PTM being investigated.
- Invert stock reagent tubes containing Ub affinity beads, pY affinity beads, SUMO 2/3 affinity beads, and Ac affinity beads several times to make sure that the beads are completely resuspended in the tube.
- For each IP assay, aliquot bead suspension into separate 1.5 mL microcentrifuge tubes on ice (IP tube). Here, add 20 µL of ubiquitin affinity beads, 30 µL of phosphotyrosine affinity beads, 40 µL of SUMOylation 2/3 affinity beads, or 50 µL of acetylation affinity beads to individual 1.5 mL microcentrifuge tubes on ice.
NOTE: See Table 4 for the recommended volume of bead suspension to use for each affinity bead. - Invert stock reagent tubes containing ubiquitination IP control beads and IgG control beads several times to make sure that the beads are completely resuspended in the tube.
- Aliquot control beads per control reaction to determine non-specific binding (Control IP tube). Here, add 20 µL of Ub control beads, 30 µL of pY control beads, 40 µL of SUMO 2/3 control beads, or 50 µL of Ac control beads to individual 1.5 mL microcentrifuge tubes on ice.
NOTE: See Table 4 for the recommended volume of bead suspension to use for each type of affinity bead. - Save a small amount of lysate (20 µL) to run as a western blot input lysate control. Add 5 µL of 5x sample buffer and boil at 95 °C for 5 min.
NOTE: See Table of Materials for buffer composition. - Add lysate to each IP tube and control IP tube. Here, 1 mg of lysate was used per IP and control reaction, resulting in a total of 8 IP reactions.
NOTE: 1.0 mg of lysate per assay is recommended as a starting point. The amount of lysate required will vary depending upon the abundance of modified target protein. - Incubate the tubes on an end-over-end rotating platform at 4 °C for 2 h. Here, an ATR tube rotator was used at speed 22.
- Collect beads by centrifugation at 3,000 – 5,000 x g for 1 min at 4 °C.
- Aspirate off as much supernatant as possible without disturbing the beads.
- Wash beads in 1 mL blastR Wash Buffer (inhibitors are not necessary at this stage) for 5 min on a 4 °C rotating platform.
- Collect beads by centrifugation at 3,000 – 5,000 x g for 1 min at 4 °C.
- Aspirate off as much supernatant as possible without disturbing the beads.
- Repeat the wash step (steps 3.10 – 3.12) two more times.
- After the final wash, completely remove buffer supernatant. Minimal disruption of the bead pellet is acceptable (up to a 5% loss). Remove residual supernatant using a fine bore protein loading tip.
- Add 30 µL of bead elution buffer, and resuspend the beads by gently tapping/flicking the side of the tube. DO NOT use a pipette at this stage.
NOTE: See Table of Materials for buffer composition. - Incubate at room temperature for 5 min.
- Gently transfer each bead suspension to a 1.5 mL microcentrifuge spin column that has been placed in a 1.5 mL microcentrifuge tube.
NOTE: It is recommended to snip the end off of the transfer pipette tip for gentler transfer. - Centrifuge at 9,000 – 10,000 x g for 1 min at room temperature to collect the IP sample.
- Add 2 µL of 2-mercaptoethanol to each sample and mix well.
NOTE: It is convenient to snap the lid off the spin column and use this to cap the collection tube for further processing. - Place samples in a 95 °C water bath for 5 min. Collect sample by centrifugation at 10,000 x g for 1 min at RT.
- If necessary, store samples at -70 °C and stop here, or proceed to running SDS-PAGE, transfer, and western blot analysis (Section 4).
4. Western Blot Analysis: Identification of Protein of Interest
- Separate samples by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transfer to a polyvinylidene fluoride (PVDF) membrane, according to standard laboratory protocols32.
- Use a primary antibody to detect the post-translationally modified versions of the protein of interest. Here, PD-L1 antibody was used to detect post-translationally modified PD-L1.
- Use ultrasensitive chemiluminescence detection reagent for detection.
NOTE: The chemiluminescent reagent should be used in conjunction with an HRP-labeled secondary antibody capable of detecting the primary antibody. - Use a volume of 2 mL of chemiluminescent reagent per mini-gel-sized transfer membrane (approximately 8 x 7 cm).
- After incubation with appropriate secondary antibody (60 min at RT is recommended), wash the blot 6 x 10 min in 30 mL of tris-buffered saline with tween-20 (TBST).
NOTE: See Table of Materials for buffer composition. - Immediately before use, mix 1 mL of chemiluminescent reagent A with 1 mL of chemiluminescent reagent B (sufficient for one 8 cm x 7 cm membrane).
- Add chemiluminescent reagent to membrane and incubate with gentle rocking at RT for 1 – 5 min prior to visualization of protein signal using x-ray film or charge-coupled device (CCD) camera imaging.
NOTE: Shorter incubation times in the chemiluminescent reagent may be necessary for highly abundant proteins.
- After incubation with appropriate secondary antibody (60 min at RT is recommended), wash the blot 6 x 10 min in 30 mL of tris-buffered saline with tween-20 (TBST).
Representative Results
The ability of these tools to investigate PTM crosstalk by effectively detecting these 4 PTMs is partly dependent on the unique lysis buffer system. The blastR denaturing buffer effectively isolates proteins from all cellular compartments similarly to other denaturing buffers, which ensures a complete protein profile (Figure 2A). Additionally, it preserves highly labile PTM signals like SUMO 2/3, which is rapidly diminished in non-denaturing buffers like radioimmunoprecipitation assay (RIPA) (Figure 2B). Importantly, when diluted appropriately, it does not affect the integrity of the affinity reagents like other denaturing buffers (e.g., Laemmli buffer).
A significant hurdle when working with denaturing buffers is the ability to effectively remove genomic DNA contamination. The conventional methodology to reduce viscosity is to shear the DNA by using a syringe needle or sonicating the sample. Figure 3A shows genomic DNA contamination in A431 cell lysate after treatment with the BlastR filter, syringe needle, or sonication. There is nearly complete removal of genomic DNA using the BlastR filter, which is not the case using a conventional syringe needle or sonication. Genomic DNA contamination significantly affects protein migration through an SDS- acrylamide gel; however, treatment with the BlastR filter removes genomic DNA, resulting in proper protein migration (Figure 3B). Altered migration caused by genomic DNA contamination can significantly affect interpretation of western blots; for example, the smeared EGFR pattern seen in the unfiltered lysate may be inaccurately interpreted as increased expression relative to the filtered sample (Figure 3C).
By utilizing this optimized PTM enrichment and detection system, one can rapidly determine if a target protein is modified by one or more PTMs. Investigation of the PD-L1 PTM profile was recently performed using this technique, and the results showed that PD-L1 was ubiquitinated, acetylated, and tyrosine phosphorylated in response to EGF (Figure 4). Importantly, these data reported endogenous PD-L1 PTM changes, which represented a small percentage of the total PD-L1 identified.

Figure 1: Filtering genomic DNA from cell lysate with BlastR filter. (A) Image of BlastR filter. (B) Lysate is loaded into the filter that was placed in a 15 mL collection tube. (C) Plunger is placed into the syringe and lysate is passed through the filter by compression. (D) Collect lysate, including any bubbles through complete compression. (E) Filtered lysate.
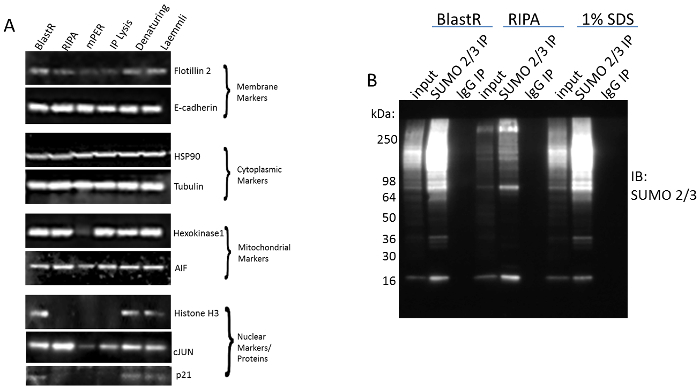
Figure 2: Comparison of BlastR lysis buffer to alternative lysis buffers. Figure 2A adapted from Horita et al. 2017. Biosci. Rep.31 (A) A431 cells were lysed with BlastR, RIPA, mPER, IP lysis, Denaturing (1% SDS), and Laemmli lysis buffers. All denaturing lysates had genomic DNA removed using BlastR filter. Isolation of proteins from the membrane, cytoplasmic, mitochondrial, and nuclear markers were determined using antibodies against the respective compartment marker proteins. (B) A431 cells were lysed with BlastR, RIPA, and 1% SDS denaturing buffer. Lysates were immunoprecipitated with SUMO 2/3 or control IgG affinity beads. Samples were separated by SDS-PAGE and analyzed by western blot using a SUMO 2/3- horseradish peroxidase (HRP) antibody. Representative blots from N≥3 independent experiments are shown. Please click here to view a larger version of this figure.
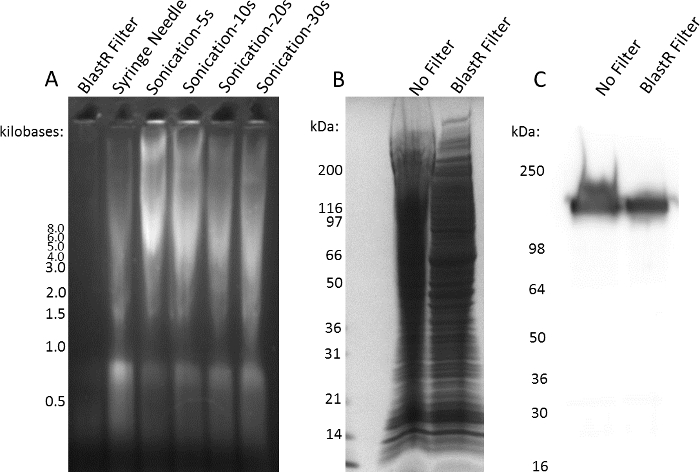
Figure 3: BlastR lysis filter is effective at removing genomic DNA. (A) A431 cells were lysed with a denaturing lysis buffer. Genomic DNA was removed or sheared with BlastR filter, syringe needle, or sonication for 5, 10, 20, or 30 seconds. 2% of the lysate was analyzed by ethidium bromide, agarose gel electrophoresis. (B) Lysate from A431 cells lysed with a denaturing buffer was either unfiltered or filter with the BlastR filter. Samples were separated with SDS-PAGE and visualized using Coomassie stain. (C) Duplicate samples from B were separated by SDS-PAGE, transferred to PVDF, and EGFR protein was examined using an EGFR antibody. Representative blots from N ≥3 independent experiments are shown. Please click here to view a larger version of this figure.
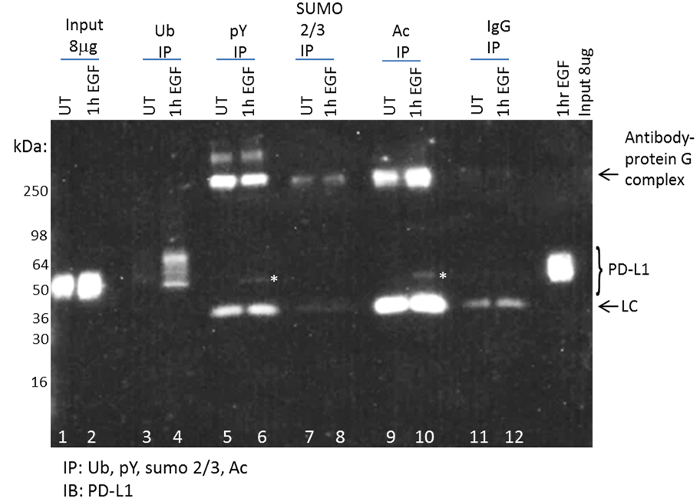
Figure 4: Detection of EGF-induced post-translational modifications for PD-L1. Figure adapted from Horita et al. 2017. Neoplasia30 (A). Serum-restricted A431 cells were either unstimulated (UT) or stimulated with EGF for one hour prior to lysis with BlastR lysis buffer. WCL was analyzed for PD-L1 levels (lanes 1,2). Ubiquitin binding beads (UBA01) were used to IP ubiquitinated proteins (lanes 3,4). Phosphotyrosine binding beads (APY03) were used to IP tyrosine-phosphorylated proteins (lanes 5,6). SUMO 2/3 binding beads (ASM24) were used to IP SUMOylated 2/3 proteins (lanes 7,8). Acetyl lysine binding beads (AAC01) were used to IP acetylated proteins (lanes 9,10). IgG binding control beads were used to IP non-specific binding proteins (lanes 11,12). Samples were separated by SDS-PAGE and analyzed by western blot using a PD-L1 antibody. A representative blot from N ≥3 independent experiments is shown. White asterisks were used to highlight PD-L1 pY and Ac protein bands. Please click here to view a larger version of this figure.
| Calculations for BlastR lysis buffer | ||||
| 1.0 mL | 2.0 mL | 5.0 mL | 10.0 mL | |
| BlastR Lysis buffer | 965 µL | 1930 µL | 4.825 mL | 9.650 mL |
| Tyrosine phosphatase Inhibitor | 5 µL | 10 µL | 25 µL | 50 µL |
| de-ubiquitinase/de-sumoylase inhibitor | 10 µL | 20 µL | 50 µL | 100 µL |
| HDAC inhibitor | 10 µL | 20 µL | 50 µL | 100 µL |
| Protease inhibitor cocktail | 10 µL | 20 µL | 50 µL | 100 µL |
| Calculations for BlastR dilution buffer | ||||
| 4.0 mL | 8.0 mL | 20.0 mL | 40.0 mL | |
| BlastR Lysis buffer | 3.86 mL | 7.72 mL | 19.3 mL | 38.6 mL |
| Tyrosine phosphatase Inhibitor | 20 µL | 40 µL | 100 µL | 200 µL |
| de-ubiquitinase/de-sumoylase inhibitor | 40 µL | 80 µL | 200 µL | 400 µL |
| HDAC inhibitor | 40 µL | 80 µL | 200 µL | 400 µL |
| Protease inhibitor cocktail | 40 µL | 80 µL | 200 µL | 400 µL |
Table 1: Lysis and Dilution Buffer Preparation Chart. Chart providing concentrations of inhibitors to add for a given lysis and dilution buffer volume when preparing buffers for cell lysis.
| Plate Protein content | Recommended BlastR Lysis Buffer volume | Recommended BlastR Dilution Buffer volume |
| < 1 mg | Combine protein from multiple plates | To make 1.5 mL final volume |
| 1-2 mg | 300 µL | To make 1.5 mL final volume |
| 2-4 mg | 600 µL | To make 3 mL final volume |
| 4-6 mg | 900 µL | To make 4.5 mL final volume |
Table 2: BlastR Lysis/Dilution Buffer Chart. Chart providing recommended lysis and dilution buffer volumes when obtaining lysate.
| Volume of cell lysate added to 1 ml of Precision Red Protein Assay reagent (µL) | Multiplier to use with sample reading OD600 |
| 10 | 10 |
| 20 | 5 |
| 30 | 3.3 |
| 40 | 2.5 |
| 50 | 2 |
Table 3: Multipliers to Convert Spectrophotometer Readings to mg/mL Lysate. Chart providing conversion numbers to aide with calculating protein concentration.
| affinity beads | volume of bead slurry/IP (mL) |
| Ubiquitination | 20 |
| Phosphotyrosine | 30 |
| SUMOylation 2/3 | 40 |
| Acetylation | 50 |
| control beads | volume of bead slurry/IP (mL) |
| Ubiquitination control beads | 20 |
| Phosphotyrsoine control beads | 30 |
| SUMOylation 2/3 control beads | 40 |
| Acetylation control beads | 50 |
Table 4: Recommended Volume of Beads/IP Chart. Chart providing recommended amounts of affinity beads to use per IP reaction.
Discussion
Initial approaches used to determine if a target protein is modified by a PTM can be performed using the target protein specific antibody for IP, followed by western blot with a PTM antibody (e.g., anti-acetyl lysine), or by using a PTM antibody for IP, followed by western blot with the target protein specific antibody30,31,33. While both approaches theoretically work, utilizing a target-protein-specific antibody has more potential pitfalls, such as the antibody may not be IP compatible, or large PTM modifications may block the antibody recognition site on the target protein34,35. The advantage of the PTM affinity beads is that the antibody or binding domains specifically recognize the PTM of interest; thus, modifications to the target protein should not alter recognition by the affinity beads. As an example, PD-L1 Ub was identified with the technique described here, and both endogenous mono- and poly-Ub was observed (Figure 4). A recent publication by Lim et al. utilized in vitro Ub techniques to investigate PD-L1 Ub, and the result was very similar to the results shown in Figure 436. Interestingly, they also performed IP with a PD-L1 antibody to enrich PD-L1 from cell lysate, where Ub was overexpressed and MG-132 was added to enhance the signal. The Ub pattern was not robust and very distinct from the in vitro Ub pattern. Investigation of endogenous PD-L1 Ub in cell culture models was not performed in the Lim et al. report to clarify the difference between their cell culture and in vitro data.
Investigating whether a protein is modified by a PTM can be challenging, due to its low abundance and transient nature37,38, and often requires enrichment through IP. Effective IP of PTMs requires optimization of several key steps and reagents, such as lysis buffers and affinity reagents. When investigating multiple PTMs of a target protein, the required optimization likely increases. Utilizing the blastR lysis system is a critical step in this protocol, as it maintains robust IP capability, while enabling PTM detection of the pY, SUMO 2/3, Ub, and Ac PTMs in a single system. This technique optimizes the time and resources required to determine if a specific target protein is modified by these four PTMs, and potentially provides a better picture of PTM crosstalk relative to comparing PTM results performed using multiple lysis systems. Investigation of the blastR lysis system's compatibility with alternative PTMs, like glycosylation, was performed; however, it has not been examined exhaustively for all types of PTMs.
Copious genomic DNA can interfere with protein measurements using either colorimetric or nanodrop methods, affect the migration of proteins in an SDS acrylamide gel, and prevent protein and affinity matrix interaction during IP assays. The method described here utilizes a specialized filter to effectively remove genomic DNA contamination, which is another critical step of this protocol. To highlight this point, viscosity tests were performed before and after blastR filter treatment, and the results showed a reduction from a high viscosity to the viscosity of water (data not shown). This change in viscosity was supported by the results in Figure 3, showing nearly all the genomic DNA had been removed. Importantly, DNA is not sheared with this method, as any sheared DNA will not be captured by the filter (data not shown). This tool is superior to conventional methods, like sonication or syringe-DNA-shearing, because it requires no specialized equipment, is highly reproducible, and removes the DNA instead of shearing it. Furthermore, it will not degrade protein in the lysate, which can occur using conventional methods29. Utilizing the filter takes 5 – 30 seconds per sample compared to alternative methods where effective breakdown of DNA may take several minutes (i.e., syringe shearing) and can result in sample heterogeneity as DNA contaminants remain in the lysate. Extensive analysis of the lysate pre- and post- blastR filtering was performed, and no observable difference in the protein profile was observed by Coomassie, target-specific western, or total and target-specific PTM analyses; thus, the integrity of the protein profile may not have been affected by filtering out the genomic DNA. Ultimately, this filter system is beneficial for any western or IP application where genomic DNA is present and may affect interpretation of the protein analysis.
It is important to note that there is potential for false negative detection utilizing this technique, which may be due in part to affinity bead saturation, binding site interference, or PTM masking. For instance, a particular target protein may be modified by Ac at very low levels; thus, it may not be isolated by pan-acetyl lysine affinity beads that have been saturated by more abundant Ac-modified target proteins. Ongoing studies are being performed to assess the detection limits of the affinity reagents utilized in this protocol, but a recent publication suggests a very robust detection limit. The data showed that this technique could identify as few as 17 acetylated target protein molecules per cell31. Still, specific circumstances such as cell type specificity, transient PTMs in response to specific stimuli, or masking of PTMs due to protein interaction may all result in false negative results. These are potential pitfalls of this technique, as well as most PTM IP methods. Thus, it is recommended to confirm results using multiple approaches.
Modifications like glycosylation and phosphorylation have been shown to compete for similar amino acids39,40, and other PTMs like ubiquitination and phosphorylation have been shown to work sequentially to regulate a protein's function17,41. Recent work on the neuropathological protein Tau highlighted the importance of PTM crosstalk, where Tau hyper-phosphorylation and Tau SUMOylation enhanced each other42. Moreover, this group showed that SUMOylation of Tau prevented poly-Ub and subsequent Tau degradation, possibly leading to aggregation. This is just one of many examples of regulatory PTM crosstalk, and the utility of this technique will help to illuminate the importance of PTMs and their crosstalk in regulating key proteins in health and disease.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank Cytoskeleton Inc. Research Scientists and Drs. Brian Hoover and Ashley Davis for their critical review, editing, and fruitful discussions on the manuscript. Additionally, we thank Dr. Robert Hom for his contribution during production of the video manuscript. This work was supported by funding from Cytoskeleton Inc.
Materials
| Cell lysis/genomic DNA removal and analysis | |||
| 1x phosphate buffered saline (PBS) | common buffer | Recipe: NaCl, KCl, Na2HPO4, KH2PO4 | |
| BlastR lysis buffer | Cytoskeleton Inc. | BLST01 | Proprietary mixture of detergents, salts, and other buffer additives |
| Radioimmunoprecipitation assay buffer (RIPA) | common buffer | Recipe: Tris-HCl, NaCl, SDS, Sodium Deoxycholate, Igepal | |
| Mammalian protein extraction reagent (mPER) | Thermofisher | 78503 | Proprietary mixture of detergents, salts, and other buffer additives |
| Immunoprecipitation (IP) Lysis | Pierce | 87787 | Proprietary mixture of detergents, salts, and other buffer additives |
| 1% sodium dodecyl sulfate (SDS) denaturing buffer | common buffer | Recipe: PBS, SDS, EDTA, EGTA | |
| Laemmli | common buffer | Recipe: Tris-HCl, SDS, glycerol, bromophenol blue | |
| BlastR dilution buffer | Cytoskeleton Inc. | BDB01 | Proprietary mixture of detergents, salts, and other buffer additives |
| Protease Inhibitor Cocktail | Cytoskeleton Inc. | PIC02 | |
| N-Ethylmaleimide (NEM) & N,N,N′,N′-Tetrakis(2-pyridylmethyl)ethylenediamine (TPEN) | Cytoskeleton Inc. | NEM09BB | |
| Trichostatin A (TSA) & Nicotinamide | Cytoskeleton Inc. | TSA01 | |
| Activated Sodium Orthovanadate (Na3VO4) | Cytoskeleton Inc. | PYI01 | |
| cell scrapers | Costar | 3008 | |
| BlastR Filter | Cytoskeleton Inc. | BLR02 | |
| BD Insulin syringe needle | BD | 08290-3284-11 | |
| sonicator (Branson Sonifier) | Branson | Sonifier 450 | |
| Precision Red Advanced Protein Reagent | Cytoskeleton Inc. | ADV02 | |
| 1 mL cuvettes | Fisher | 14955127 | |
| Spectrophotometer (Genesys20) | Thermofisher | 4001 | Other spectrophotomers can be used to measure protein concentration |
| Agarose | Fisher | BP1356-500 | |
| 1kb Deoxyribonucleic acid (DNA) ladder | NEB | N3232L | |
| DNA gel box | Thermofisher | 7312 B1A | |
| Tris/Borate/EDTA (TBE) buffer | common buffer | Recipe: Tris-HCl, Boric Acid, EDTA | |
| PTM Immunoprecipitation | |||
| Phosphotyrosine beads | Cytoskeleton Inc. | APY03-beads | |
| Ubiquitination beads | Cytoskeleton Inc. | UBA01-beads | |
| SUMOylation 2/3 beads | Cytoskeleton Inc. | ASM24-beads | |
| Acetylation beads | Cytoskeleton Inc. | AAC04-beads | |
| Phosphotyrosine control beads | Cytoskeleton Inc. | CIG01-beads | |
| Ubiquitination control beads | Cytoskeleton Inc. | CUB02-beads | |
| SUMOylation 2/3 control beads | Cytoskeleton Inc. | CIG01-beads | |
| Acetylation control beads | Cytoskeleton Inc. | CIG02-beads | |
| BlastR wash buffer | Cytoskeleton Inc. | BWB02 | Proprietary mixture of detergents, salts, and other buffer additives |
| Bead elution buffer | Cytoskeleton Inc. | BEB01 | Proprietary mixture of detergents, salts, and other buffer additives |
| microcentrifuge | Eppendorf | 5417 | Other microcentrifuges can be used |
| tube rotator | ATR | RKVSD | Other end-over-end tube rotators can be used |
| protein loading tips | VWR | 37001-150 | |
| spin columns | Cytoskeleton Inc. | SPN22 | |
| heat block 95 °C | Benchmark | BSH1002 | |
| SDS-PAGE/Transfer/Western analysis | |||
| 5x sample buffer | common buffer | recipe: Bromophenol blue, dithiothreitol (DTT), Glycerol, SDS, Tris-HCl | |
| novex wedge well 4-20% Tris-gly gels | Invitrogen | XP04200BOX | The large wedgewells are excellent for loading large volumes |
| 1x Running buffer | common buffer | recipe: Tris-HCl , Glycine, SDS | |
| seeblue protein ladder | Life Technologies | LC5625 | |
| electrophoresis cell | Invitrogen | Xcell surelock electrophoresis cell | Other electrophoresis cells can be used, however these are compatible the novex wedgewell gels |
| power supply | Biorad | PowerPac HC | Other power supplies can be used |
| 1x Transfer buffer | common buffer | recipe: Tris-HCl, glycine, methanol | |
| Transfer cell | Biorad | mini trans-blot cell | Other transfer cells can be used |
| Immobilon- P membranes (PVDF) | millipore | IPVH 304F0 | |
| non-fat milk | thrive | 813503015095 | |
| Tris buffered saline with tween (TBST) | common buffer | recipe: Tris-HCl, NaCl, Tween 20 | |
| Chemiluminescent Detection Reagent | Cytoskeleton Inc. | CLRA/CLRB | |
| Cell growth and treatment | |||
| Dulbecco's Modified Eagle's medium | ATCC | 30-2002 | |
| Fetal bovine serum | ATLAS | F-0500-A | |
| Epidermal growth factor | Cytoskeleton Inc. | CN02-A | |
| 150 mm cell culture plates | Corning | 430599 |
References
- Drazic, A., Myklebust, L. M., Ree, R., Arnesen, T. The world of protein acetylation. Biochim Biophys Acta. 1864 (10), 1372-1401 (2016).
- Swatek, K. N., Komander, D. Ubiquitin modifications. Cell Res. 26 (4), 399-422 (2016).
- Eifler, K., Vertegaal, A. C. SUMOylation-Mediated Regulation of Cell Cycle Progression and Cancer. Trends Biochem Sci. 40 (12), 779-793 (2015).
- Hunter, T. The genesis of tyrosine phosphorylation. Cold Spring Harb Perspect Biol. 6 (5), 1-15 (2014).
- International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 431 (7011), 931-945 (2004).
- Jensen, O. N. Modification-specific proteomics: characterization of post-translational modifications by mass spectrometry. Curr Opin Chem Biol. 8 (1), 33-41 (2004).
- Pellegrino, S., Altmeyer, M. Interplay between Ubiquitin, SUMO, and Poly(ADP-Ribose) in the Cellular Response to Genotoxic Stress. Front Genet. 7 (63), 1-8 (2016).
- Liddy, K. A., White, M. Y., Cordwell, S. J. Functional decorations: post-translational modifications and heart disease delineated by targeted proteomics. Genome Med. 5 (2), 20 (2013).
- Margolin, D. H., Kousi, M., Chan, Y. M., Lim, E. T., Schmahmann, J. D., Hadjivassiliou, M., Hall, J. E., Adam, I., Dwyer, A., Plummer, L., et al. Ataxia, dementia, and hypogonadotropism caused by disordered ubiquitination. N Engl J Med. 368 (21), 1992-2003 (2013).
- Droescher, M., Chaugule, V. K., Pichler, A. SUMO rules: regulatory concepts and their implication in neurologic functions. Neuromolecular Med. 15 (4), 639-660 (2013).
- Anbalagan, M., Huderson, B., Murphy, L., Rowan, B. G. Post-translational modifications of nuclear receptors and human disease. Nucl Recept Signal. 10 (e001), 1-13 (2012).
- Kim, M. Y., Bae, J. S., Kim, T. H., Park, J. M., Ahn, Y. H. Role of transcription factor modifications in the pathogenesis of insulin resistance. Exp Diabetes Res. 2012 (716425), 1-16 (2012).
- West, A. C., Johnstone, R. W. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 124 (1), 30-39 (2014).
- Kim, H. J., Bae, S. C. Histone deacetylase inhibitors: molecular mechanisms of action and clinical trials as anti-cancer drugs. Am J Transl Res. 3 (2), 166-179 (2011).
- Gu, B., Zhu, W. G. Surf the post-translational modification network of p53 regulation. Int J Biol Sci. 8 (5), 672-684 (2012).
- Janke, C. The tubulin code: molecular components, readout mechanisms, and functions. J Cell Biol. 206 (4), 461-472 (2014).
- Nguyen, L. K., Kolch, W., Kholodenko, B. N. When ubiquitination meets phosphorylation: a systems biology perspective of EGFR/MAPK signalling. Cell Commun Signal. 11 (52), 1-15 (2013).
- Hunter, T. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol Cell. 28 (5), 730-738 (2007).
- Butler, P. L., Staruschenko, A., Snyder, P. M. Acetylation stimulates the epithelial sodium channel by reducing its ubiquitination and degradation. J Biol Chem. 290 (20), 12497-12503 (2015).
- Wang, Y., Wang, Y., Zhang, H., Gao, Y., Huang, C., Zhou, A., Zhou, Y., Li, Y. Sequential posttranslational modifications regulate PKC degradation. Mol Biol Cell. 27 (2), 410-420 (2016).
- Guo, Z., Kanjanapangka, J., Liu, N., Liu, S., Liu, C., Wu, Z., Wang, Y., Loh, T., Kowolik, C., Jamsen, J., et al. Sequential posttranslational modifications program FEN1 degradation during cell-cycle progression. Mol Cell. 47 (3), 444-456 (2012).
- Cui, W., Sun, M., Zhang, S., Shen, X., Galeva, N., Williams, T. D., Staudinger, J. L. A SUMO-acetyl switch in PXR biology. Biochim Biophys Acta. 1859 (9), 1170-1182 (2016).
- Doll, S., Burlingame, A. L. Mass spectrometry-based detection and assignment of protein posttranslational modifications. ACS Chem Biol. 10 (1), 63-71 (2015).
- Siuti, N., Kelleher, N. L. Decoding protein modifications using top-down mass spectrometry. Nat Methods. 4 (10), 817-821 (2007).
- Mischerikow, N., Heck, A. J. Targeted large-scale analysis of protein acetylation. Proteomics. 11 (4), 571-589 (2011).
- Barysch, S. V., Dittner, C., Flotho, A., Becker, J., Melchior, F. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nat Protoc. 9 (4), 896-909 (2014).
- Emmerich, C. H., Cohen, P. Optimising methods for the preservation, capture and identification of ubiquitin chains and ubiquitylated proteins by immunoblotting. Biochem Biophys Res Commun. 466 (1), 1-14 (2015).
- Peach, M., Marsh, N., Miskiewicz, E. I., MacPhee, D. J. Solubilization of proteins: the importance of lysis buffer choice. Methods Mol Biol. 1312, 49-60 (2015).
- Grabski, A. C. Advances in preparation of biological extracts for protein purification. Methods Enzymol. 463, 285-303 (2009).
- Horita, H., Law, A., Hong, S., Middleton, K. Identifying Regulatory Posttranslational Modifications of PD-L1: A Focus on Monoubiquitinaton. Neoplasia. 19 (4), 346-353 (2017).
- Horita, H., Law, A., Hong, S., Middleton, K. A simple toolset to identify endogenous post-translational modifications for a target protein: a snapshot of the EGFR signaling pathway. Biosci Rep. 37 (4), 1-14 (2017).
- Mahmood, T., Yang, P. C. Western blot: technique, theory, and trouble shooting. N Am J Med Sci. 4 (9), 429-434 (2012).
- Li, C. L., Sathyamurthy, A., Oldenborg, A., Tank, D., Ramanan, N. SRF phosphorylation by glycogen synthase kinase-3 promotes axon growth in hippocampal neurons. J Neurosci. 34 (11), 4027-4042 (2014).
- Fuchs, S. M., Strahl, B. D. Antibody recognition of histone post-translational modifications: emerging issues and future prospects. Epigenomics. 3 (3), 247-249 (2011).
- Fuchs, S. M., Krajewski, K., Baker, R. W., Miller, V. L., Strahl, B. D. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 21 (1), 53-58 (2011).
- Lim, S. O., Li, C. W., Xia, W., Cha, J. H., Chan, L. C., Wu, Y., Chang, S. S., Lin, W. C., Hsu, J. M., Hsu, Y. H., et al. Deubiquitination and Stabilization of PD-L1 by CSN5. Cancer Cell. 30 (6), 925-939 (2016).
- Wu, R., Haas, W., Dephoure, N., Huttlin, E. L., Zhai, B., Sowa, M. E., Gygi, S. P. A large-scale method to measure absolute protein phosphorylation stoichiometries. Nat Methods. 8 (8), 677-683 (2011).
- Ordureau, A., Munch, C., Harper, J. W. Quantifying ubiquitin signaling. Mol Cell. 58 (4), 660-676 (2015).
- Lefebvre, T., Ferreira, S., Dupont-Wallois, L., Bussiere, T., Dupire, M. J., Delacourte, A., Michalski, J. C., Caillet-Boudin, M. L. Evidence of a balance between phosphorylation and O-GlcNAc glycosylation of Tau proteins–a role in nuclear localization. Biochim Biophys Acta. 1619 (2), 167-176 (2003).
- Wang, X., Li, D., Wu, G., Bazer, F. W. Functional Roles of Fructose: Crosstalk between O-Linked Glycosylation and Phosphorylation of Akt-TSC2-MTOR Cell Signaling Cascade in Ovine Trophectoderm Cells. Biol Reprod. 9 (5), 1-17 (2016).
- Levkowitz, G., Waterman, H., Zamir, E., Kam, Z., Oved, S., Langdon, W. Y., Beguinot, L., Geiger, B., Yarden, Y. c-Cbl/Sli-1 regulates endocytic sorting and ubiquitination of the epidermal growth factor receptor. Genes Dev. 12 (23), 3663-3674 (1998).
- Luo, H. B., Xia, Y. Y., Shu, X. J., Liu, Z. C., Feng, Y., Liu, X. H., Yu, G., Yin, G., Xiong, Y. S., Zeng, K., et al. SUMOylation at K340 inhibits tau degradation through deregulating its phosphorylation and ubiquitination. Proc Natl Acad Sci U S A. 111 (46), 16586-16591 (2014).

