Merkel Cell Polyomavirus Infection and Detection
Summary
Here, we present a protocol to infect primary human dermal fibroblast with MCPyV. The protocol includes isolation of dermal fibroblasts, preparation of MCPyV virions, virus infection, immunofluorescence staining, and fluorescence in situ hybridization. This protocol can be extended for characterizing MCPyV-host interactions and discovering other cell types infectable by MCPyV.
Abstract
Merkel cell polyomavirus (MCPyV) infection can lead to Merkel cell carcinoma (MCC), a highly aggressive form of skin cancer. Mechanistic studies to fully investigate MCPyV molecular biology and oncogenic mechanisms have been hampered by a lack of adequate cell culture models. Here, we describe a set of protocols for performing and detecting MCPyV infection of primary human skin cells. The protocols describe the isolation of human dermal fibroblasts, preparation of recombinant MCPyV virions, and detection of virus infection by both immunofluorescent (IF) staining and in situ DNA-hybridization chain reaction (HCR), which is a highly sensitive fluorescence in situ hybridization (FISH) approach. The protocols herein can be adapted by interested researchers to identify other cell types or cell lines that support MCPyV infection. The described FISH approach could also be adapted for detecting low levels of viral DNAs present in the infected human skin.
Introduction
Merkel cell polyomavirus (MCPyV) is a small, double-stranded DNA virus that has been associated with a rare but aggressive skin cancer, Merkel cell carcinoma (MCC)1,2. The mortality rate of MCC, around 33%, exceeds that of melanoma3,4. MCPyV has a circular genome of ~5 kb1,5 bisected by a non-coding regulatory region (NCRR) into early and late coding regions1. The NCRR contains the viral origin of replication (Ori) and bidirectional promoters for viral transcription6,7. The early region encodes tumor antigen proteins called large T (LT), small T (sT), 57kT, alternative LT ORF (ALTO), as well as an autoregulatory miRNA1,8,9,10. The late region encodes the capsid proteins VP1 and VP211,12,13. LT and sT are the best-studied MCPyV proteins and have been shown to support the viral DNA replication and MCPyV-induced tumorigenesis5. Clonal integration of MCPyV DNA into the host genome, which has been observed in up to 80% of MCCs, is likely a causal factor for virus-positive tumor development14,15.
The incidence of MCC has tripled over the past twenty years16. Asymptomatic MCPyV infection is also widespread in the general population17,18,19. With the increasing number of MCC diagnoses and the high prevalence of MCPyV infection, there is a need to improve our understanding of the virus and its oncogenic potential. However, many aspects of MCPyV biology and oncogenic mechanisms remain poorly understood20. This is largely because MCPyV replicates poorly in established cell lines11,12,21,22,23 and, until recently, skin cells capable of supporting MCPyV infection had not been discovered22. Mechanistic studies to fully investigate MCPyV and its interaction with host cells have been hampered by a lack of cell culture system for propagating the virus5.
We discovered that primary human dermal fibroblasts (HDFs) isolated from neonatal human foreskin support robust MCPyV infection both in vitro and ex vivo24. From this study, we established the first cell culture infection model for MCPyV24. Building on this model system, we showed that the induction of matrix metalloproteinase (MMP) genes by the WNT/β-catenin signaling pathway and other growth factors stimulates MCPyV infection. Moreover, we found that the FDA-approved MEK antagonist trametinib effectively inhibits MCPyV infection5,25. From these studies, we also established a set of protocols for isolating human dermal fibroblasts24,25, preparing MCPyV virions11,12, performing MCPyV infection on human dermal fibroblasts24,25 and detecting MCPyV proteins by IF staining26. In addition, we adapted the in situ DNA hybridization chain reaction (HCR) technology27 to develop a highly sensitive FISH technique (HCR-DNA FISH) for detecting MCPyV DNA in infected human skin cells. These new methods will be useful for studying the infectious cycle of MCPyV as well as the cellular response to MCPyV infection. The natural host reservoir cells that maintain MCPyV infection and the cells that give rise to MCC tumors remain unknown. The techniques we describe in this manuscript could be applied to examine various types of human cells to identify both the reservoir cells and origin of MCC tumors. Our established methods, such as HCR-DNA FISH, could also be employed in the detection of other DNA tumor viruses and the characterization of host cell interactions.
Protocol
Human neonatal foreskins were obtained from Penn Skin Disease Research Center. Adult human fibroblasts were obtained from discarded normal skin after surgery. All the protocols were approved by the University of Pennsylvania Institutional Review Board.
1. Isolation of human dermal fibroblasts
- Use a pair of scissors to trim off fat and subcutaneous tissue from the human neonatal foreskin and cut the skin sample in halves or quarters.
- Incubate the tissue in 5 mL of 10 mg/mL dispase II in PBS supplemented with antibiotic-antimycotic at 4 °C overnight.
- In a sterile 10 cm dish, carefully separate the epidermis from the dermal layers using microdissection forceps.
- Transfer the resulting dermal tissue to a 15 mL conical tube containing 5 mL of 2 mg/mL collagenase type IV in FBS-free medium supplemented with antibiotic-antimycotic.
- Incubate the sample at 37 °C in 5% CO2 for 4-6 h with periodic shaking until just a few macroscopic tissue aggregates remain.
- Release single cells from the dermis by pipetting up and down (triturating) vigorously ten times with a 10 mL pipette.
- Centrifuge at 180 x g for 5 min, and discard the supernatant.
- Plate the dissociated cells in DMEM medium supplemented with 10% fetal calf serum, 1% non-essential amino acids, and 1% L-glutamine at 37 °C in 5% CO2.
2. Recombinant MCPyV virion preparation
- Digest 50 µg of pR17b plasmid (carrying MCPyV genome) with 250 U of BamHI-HF in a 200 µL volume (4 h at 37 °C) to separate the viral genome from the vector backbone (Figure 2).
- Add 1200 µL of buffer PB (supplemented with 10 µL of 3 M NaAc, pH 5.2) to the digested DNA and purify over 2 miniprep spin columns (20 µg DNA capacity). Elute the digested pR17b plasmid from each column with 200 µL of TE buffer (10 mM Tris-HCl, pH8.0, 1 mM EDTA).
- Prepare the ligation reaction in a 50 mL centrifuge tube. Add 400 µL of purified plasmid DNA from step 2.2, 8.6 mL of 1.05x T4 ligase buffer and 6 µL of high concentration T4 ligase. Incubate at 16 °C overnight.
- Add 45 mL of buffer PB (supplemented with 10 µL of 3 M NaAc, pH 5.2) to the ligation and use a vacuum manifold to load through 2 miniprep spin columns. Elute each column with 50 µL of TE buffer. Expect a yield of about 30 µg of DNA.
- In the late afternoon/evening, seed 6 x 106 293TT28 cells into a 10 cm dish containing DMEM medium supplemented with 10% fetal calf serum, 1% non-essential amino acids, and 1% L-glutamine without hygromycin B.
- The next morning, ensure that the cells are about 50% confluent. Transfect using 66 µL of transfection reagent (1.1 µL/cm2), 12 µg of re-ligated MCPyV isolate R17b DNA from step 2.4, 8.4 µg of ST expression plasmid pMtB and 9.6 µg of LT expression plasmid pADL*29.
- When the transfected cells are nearly confluent, the following day, trypsinize the cells and transfer them to a 15 cm dish for continued expansion.
- Optionally take a small number of 293TT cells upon expansion and perform IF staining for MCPyV LT (CM2B4) and VP1 (MCV VP1 rabbit30) to determine transfection efficiency. At this stage, nuclear LT signal may be visible but VP1 expression probably will not be detectable.
- When the 15 cm dish becomes nearly confluent (usually 5-6 days after initial transfection), transfer the cells into three 15 cm dishes. Harvest the cells from the 15 cm dishes when they become nearly confluent and follow the virus harvest protocol below.
NOTE: Optionally perform quality control IF as described in step 2.8. Most of the cells should be both MCPyV LT and VP1 positive at this stage. - To harvest the virus, trypsinize cells, spin at 180 x g for 5 min at RT and remove the supernatant. Add one cell pellet volume of DPBS-Mg (DPBS with 9.5 mM MgCl2 and 1x antibiotic-antimycotic). Then add 25 mM ammonium sulfate (from a 1 M pH 9 stock solution) followed by 0.5% Triton X-100 (from a 10% stock solution), 0.1% Benzonase and 0.1% of an ATP-Dependent DNase. Mix well and incubate at 37 °C overnight.
- Incubate the mixture for 15 min on ice and then add 0.17 volume of 5M NaCl. Mix and incubate on ice for another 15 min. Spin for 10 min at 12,000 x g in a 4 °C centrifuge. If the supernatant is not clear, gently invert the tube and repeat the spinning step. Transfer the supernatant to a new tube.
- Resuspend the pellet using one volume of DPBS supplemented with 0.8 M NaCl, and spin again, as described in step 2.11.
- Combine the supernatants from the step 2.11 and 2.12, and spin one more time as described in step 2.11.
- Pour gradients of iodixanol in thin wall 5 mL polyallomer tubes by underlaying (27%, then 33%, then 39%) ~0.7 mL steps using a 3 mL syringe fitted with a long needle or a p1000 pipette.
- Load 3 mL of clarified virus-containing supernatant on the prepared iodixanol gradient.
- Spin for 3.5 h at 234,000 x g and 16 °C in an SW55ti rotor. Set the acceleration and deceleration to slow.
- After ultracentrifugation, collect 12 fractions in siliconized tubes (each fraction is ~400 µL).
- Analyze the fractions for the presence of virus by dsDNA reagent and/or Western blot for VP1 (MCV VP1 rabbit30). Pool gradient fractions with peak dsDNA and/or VP1 content and characterize the stock by quantitative PCR to calculate the viral genome equivalent31. Store MCPyV virion stock at -80 °C.
3. Infection
- Maintain primary human dermal fibroblasts in DMEM with 10% fetal calf serum, 1% non-essential amino acids and 1% L-glutamine. Upon reaching confluence, split fibroblasts 1:4 without spinning down.
NOTE: For highest MCPyV infection efficiency, use primary fibroblasts between passages 5 and 12 that are actively dividing at the time of plating. - To infect human dermal fibroblasts, aspirate the medium and wash the cells with DPBS.
- Add 1 mL of 0.05% Trypsin-EDTA to the dish and incubate at 37 °C for 5-10 min.
- Check under the microscope to make sure that the cells are coming off the dish.
- Add 10 mL of DMEM/F12 medium containing 20 ng/mL EGF, 20 ng/mL bFGF and 3 µM CHIR99021, all of which stimulate MCPyV infection24, to the dish and transfer the cell solution into a 15 mL tube.
- Spin down the cells at 180 x g for 2 min, and discard the supernatant. Resuspend the cells in DMEM/F12 medium containing 20 ng/mL EGF, 20 ng/mL bFGF and 3µM CHIR99021 at 2-4 x 104 cells per mL.
- Seed 200 µL of the cell suspension supplemented with 1 mg/mL collagenase type IV into each well of a 96-well plate. Thaw MCPyV virion stock on ice. Add 109 viral genome equivalents of MCPyV virions (calculated in step 2.18) per 1 µL of iodixanol for each 2,500 to 5000 cells to be infected. Tap the side of the plate gently and place the plate in the incubator.
- After incubation at 37 °C in 5% CO2 for 48-72 h, add 20% FBS to each well.
- Allow the infection to proceed at 37 °C in 5% CO2 for 72-96 h.
4. Immunofluorescent staining
- Fix cells obtained in step 3.9 with 3% paraformaldehyde in PBS for 20 min.
- Wash the cells with PBS twice.
- Incubate the cells in blocking/permeabilization buffer (0.5% Triton X-100, 3% BSA in PBS filtered through 0.22 µm filter) at RT for 1 h.
- Incubate the cells with the following primary antibodies: mouse monoclonal anti-MCPyV LT (CM2B4) (1:1000) and rabbit polyclonal anti-MCPyV VP1 antibody (1:2000), at RT for 3 h.
- Wash the cells with PBS three times.
- Incubate the cells with the secondary antibodies: Fluor 594 goat anti-mouse IgG (1:1000) and Fluor 488 goat anti-rabbit IgG (1:500) at RT for 1 h.
- Wash cells with PBS three times.
- Counterstain the cells with 0.5 µg/mL DAPI (4',6-Diamidino-2-Phenylindole, Dihydrochloride).
- Mount coverslips on glass slides and analyze using an inverted fluorescence microscope.
5. In situ DNA-HCR
NOTE: This technique requires that cells be seeded on coverslips. For this purpose, the infection conditions described above (step 3) may be scaled up to the 24-well plate format.
- Fix cells cultured on coverslips with 4% PFA in PBS for 10 min. Wash the coverslips twice with PBS, then treat them with 70% ethanol to permeabilize the cells at 4 °C overnight.
NOTE: Fixed samples can remain in this state for several days. - Pre-hybridize samples by replacing the ethanol with probe hybridization buffer and incubating for 60 min at RT.
- Dilute the probe(s) (1 µM) (Table 1) in probe hybridization buffer solution at 1:500 dilution 30 min prior to the end of the pre-hybridization step and incubate at 45 °C.
- At the end of the incubations, pipette ~10 μL of the diluted probe mix per coverslip on microscope slides. Place the coverslips, cell-side down, on their respective droplets of hybridization probe mix. Seal the edges and backs of the coverslips to the slides with a liberal amount of rubber cement.
- Heat the slides with added probes at 94 °C for 3 min by setting the slides on the flat side of a heat block. Transfer the slides to a humidified chamber and incubate at 45 °C overnight. To make a simple humidified chamber, lightly dampen sterile paper towels with dH2O and place in the bottom of a plastic container that seals with a rubber gasket.
NOTE: During heating the rubber cement can bubble briefly. However, ensure that the seal does not break. - Carefully peel away the rubber cement with forceps and place the coverslips cell-side up back into wells of a 24-well plate. Wash the coverslips with probe wash buffer at RT three times.
- Incubate the coverslip in the amplification buffer (200 µL in 24-well plates) at RT 30-60 min.
- Meanwhile, anneal each of the two labeled oligonucleotide hairpins recognizing the probes (Table 1) in separate PCR tubes by heating to 94 °C for 90 s and cooling to RT for 30 min while protecting from light. Mix the two hairpins in amplification buffer, each at a dilution of 1:50.
- Make a surface for the amplification reaction by stretching paraffin film over the open face of a 24-well plate lid. Pipette 50-100 µL droplets of the hairpin/amplification buffer mixture onto the paraffin film for each coverslip. Use forceps to carefully remove the coverslips from the pre-amplification solution. Dry the coverslip by touching the edge to a porous disposable wipe, and place cell-side down onto the amplification droplet.
- Place the plate lid holding the coverslips into a humidified chamber and incubate overnight at RT in the dark.
- Return the coverslips to wells once more. Incubate the samples with 5x SSCT [5x sodium chloride sodium citrate (SSC) 0.1% Tween 20 filtered through a 0.22 µm filter] containing 0.5 µg/mL DAPI for 1 h at RT. Wash the samples twice with 5x SSCT at RT.
- Mount the coverslips on microscope slides. Analyze the cells and image the samples using an inverted fluorescence microscope.
Representative Results
The protocol described in this manuscript allowed isolation of a nearly homogenous population of HDFs (Figure 1). As demonstrated by immunofluorescent staining, almost 100% of the human dermal cells isolated using the conditions described in this protocol were positively stained for dermal fibroblast markers, vimentin, and collagen I24, but negative for human foreskin keratinocyte marker K14 (Figure 1). Figure 2 shows the process of generating MCPyV virions using recombinant MCPyV genome and a virion sample after ultracentrifuge concentration. After visualizing the band of MCPyV virions concentrated in the core of the gradient (marked by an arrow in Figure 2B), 500 µL fractions were collected and MCPyV qPCR was performed to identify the peak fractions. An example of the qPCR analysis data is shown in Figure 2C. In some other experiments, the samples were also analyzed with Western blotting using a rabbit-anti-MCPyV VLP antibody (Supplemental Figure 1). Figure 3 shows immunofluorescent stained images of HDFs infected with MCPyV. To manually quantify positive cells, we counted at least three random views of about 300 cells. We consider cells that are LT positive, VP1 positive, or both LT and VP1 positive to be positive for MCPyV infection. In the experiments shown in Figure 3, over 30% of cells are LT positive and more than 10% are VP1 positive. The MCPyV genomes replicated in the infected cells were detected using both the HCR-DNA FISH (Figure 4A) and Immunofluorescent-HCR-DNA FISH (Figure 4B). While the HCR-DNA FISH reveals the localization of MCPyV DNA present in the replication factory (foci) in Figure 4A, the Immunofluorescent-HCR-DNA FISH methods allows simultaneous detection of both MCPyV DNA and LT protein co-localizing at the replication centers (Figure 4B). The images from the immunofluorescent-HCR-DNA FISH demonstrated that this technique could be used to reveal co-localization of viral DNA and the associated viral protein.
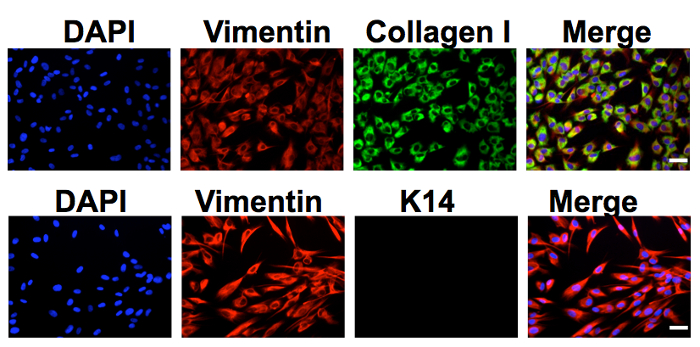
Figure 1. Human dermal fibroblasts isolated from neonatal human foreskin. Human dermal cells cultured in DMEM/F12 medium supplemented with 10% FBS were stained using antibodies against vimentin, collagen I, or keratin 14 (K14). The cells were also counterstained with DAPI. Bar, 50 µm. This figure was adapted from Figure S2 of Liu et al., 201624. Please click here to view a larger version of this figure.
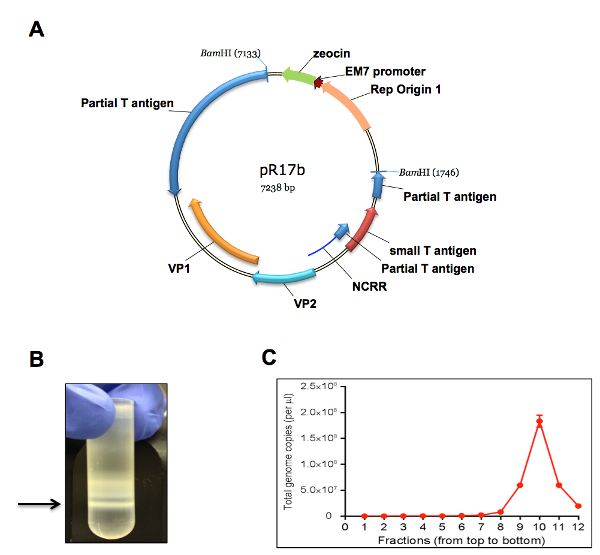
Figure 2. Production of MCPyV virion using recombinant viral genome. (A) A plasmid map of pR17b (MCPyV genome plasmid). (B) A representative picture of an MCPyV virion sample harvested and purified over a gradient. Arrow marks the band of MCPyV virions concentrated in the core of the gradient. (C) The viral genome copy number in each gradient fraction was quantified using qPCR. Core gradient fractions (numbers, counting from the top of the gradient, are indicated at the bottom of the graph). Error bars represent standard error of the mean (S.E.M.) of three technical repeats. Please click here to view a larger version of this figure.
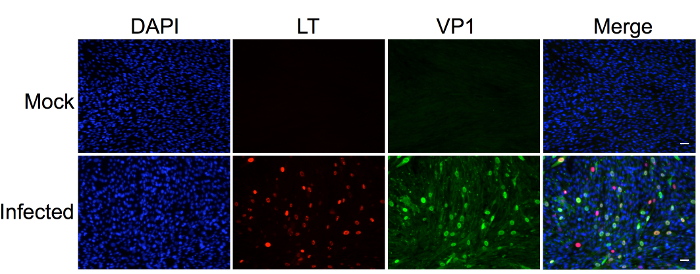
Figure 3. Human dermal fibroblasts support robust MCPyV infection, transcription, and replication. Dermal fibroblasts isolated from human skin were treated with MCPyV virions in DMEM F12 medium containing EGF, bFGF, CHIR99021, and collagenase IV for two days. After changing to fresh DMEM/F12 medium containing 20% FBS for three more days, cells were immuno-stained using the indicated antibodies and counterstained with DAPI. Many of cells not only have highly expressed LT and VP1 but also show robust MCPyV replication foci. This figure was adapted from Figure 3 of Liu et al., 201624. Scale bar, 50 m. Please click here to view a larger version of this figure.
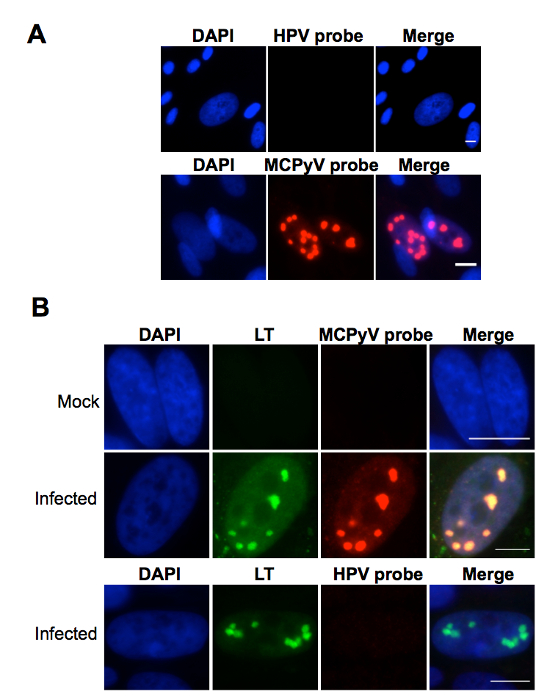
Figure 4. Detection of MCPyV in infected cells using FISH techniques. (A) Human dermal cells cultured in DMEM/F12 containing EGF, bFGF, CHIR99021, and collagenase type IV were treated with MCPyV for 2 days. After changing to fresh DMEM/F12 containing FBS for 3 more days, the cells were subjected to HCR-DNA FISH analysis. The cells were also counterstained with DAPI. (B) Human dermal cells cultured in medium containing EGF, bFGF, and CHIR99021 were either untreated (Mock) or treated (Infected) with MCPyV as described in (A). The cells were subjected to immuno-FISH using LT antibody and MCPyV-specific DNA probes before counterstaining with DAPI. HPV16 probes were used as a negative control. Scale bar, 10 m. This figure was adapted from Figure 4 of Liu et al., 201624. Please click here to view a larger version of this figure.
| Probe name | Sequences |
| MCPyV probe 1 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA tgagctacctcactaaggagtggtttttatactgcagtttcccgcccttg ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 2 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA agaggcctcggaggctaggagccccaagcctctgccaacttgaaaaaaaa ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 3 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA cattgactcatttcctggagaggcggagtttgactgataaacaaaacttt ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 4 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA gatactgccttttttgctaattaagcctcttaagcctcagagtcctctct ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 5 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA aagcttctcctgtaagaatagcttccaaagttactcctgtggtggcactt ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 6 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA ggatgttgccataacaattaggagcaatctccaaaagcttgcacagagcc ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 7 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA gctcaggggaggaaagtgattcatcgcagaagagatcctcccaggtgcca ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 8 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA aagcctgggacgctgagaaggacccatacccagaggaagagctctggctg ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 9 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA agcttcgggaccccccaaattttcgctttcttgagaatggaggaggggtc ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| MCPyV probe 10 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA cttttggctagaacagtgtctgcggcttgttggcaaatggttttctgaga ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| HPV probe 1 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA cagctctgtgcataactgtggtaactttctgggtcgctcctgtgggtcct ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| HPV probe 2 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA acaatattgtaatgggctctgtccggttctgcttgtccagctggaccatc ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| HPV probe 3 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA gtcagctatactgggtgtaagtccaaatgcagcaatacaccaatcgcaac ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| HPV probe 4 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA ctttggtatgggtcgcggcggggtggttggccaagtgctgcctaataatt ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
| HPV probe 5 | CCTCAACCTACCTCCAACTCTCACCATATTCGCTTC TAAAA ccatccattacatcccgtaccctcttccccattggtacctgcaggatcag ATTTT CACATTTACAGACCTCAACCTACCTCCAACTCTCAC |
Table 1: Probes used in the study.
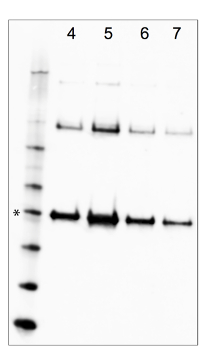
Supplemental Figure 1. Screening Iodixanol gradient fractions for MCPyV VP1. MCPyV-infected cells were lysed and subjected to Iodixanol gradient fractionation. Core gradient fractions (numbers, counting from the bottom of the gradient in this experiment, are indicated at the top of the figure) were subjected to Western blot analysis to detect VP1. 10 µL samples of each fraction were adjusted to 1x load dye with a 5% final concentration of 2-mercaptoethanol and briefly heated to 65 °C. The samples were separated on a 4-12% bis-Tris gel and blotted onto nitrocellulose. Western blotting was conducted in TBST with nonfat dry milk using a rabbit-anti-MCPyV VLP antiserum diluted 1:5000. The antiserum is available upon request. The 50 kDa standard (which migrates similarly to the 47 kDa MCPyV VP1 monomer) is marked with an asterisk. In the shown image, sample reduction was incomplete and disulfide-linked VP1 multimers are apparent. Use of dithiothreitol and/or higher denaturation temperatures can result in more complete reduction to the VP1 monomer species. Please click here to view a larger version of this figure.
Discussion
The methods described above , including isolation of dermal fibroblasts from human skin tissue, preparation of recombinant MCPyV virions, infection of cultured cells, immunofluorescent staining, and a sensitive FISH method adapted from HCR technology, which should enable researchers to analyze MCPyV infection27. One of the most critical steps to achieving MCPyV infection in vitro is the production of high-titer virion preparations. Using the protocol for preparation of recombinant MCPyV virions described here, we achieve viral titers of 1012 genome copies per mL of idoxanol solvent11,12. These high-titer MCPyV preparations allowed us to identify HDFs as the only primary skin cell type thus far that appears to support the full MCPyV infection cycle24.
Isolating fresh human dermal fibroblasts using the protocol described in this manuscript has enabled us to control the quality of the cells used in MCPyV infection experiment and ensure consistency of findings across cells isolated from different patient samples. All human dermal fibroblasts isolated from neonatal skin should be assessed for presence of MCPyV DNA by qPCR to ensure the absence of MCPyV infection prior to treatment with recombinant MCPyV virions. In order to obtain the maximum MCPyV infection efficiency, it is critical to use human dermal fibroblasts isolated freshly from neonatal human foreskin as higher passage fibroblasts support lower MCPyV infection efficiencies. Further, when isolated in parallel from adult skin samples, fibroblasts from neonatal skin samples supported much higher MCPyV infection efficiency24. We have never tested any commercially available cells for MCPyV infection. However, we see no specific reason that commercially isolated fibroblasts should be inferior other than if they are higher passage at the point of freezing.
We have also used the protocol described here to isolate dermal fibroblasts from other animals. For example, we have used the protocol to successfully isolate dermal fibroblasts from mouse (Mus musculus), rat (Rattus norvegicus), tree shrew (Tupaia Belangeri), and rhesus macaque (Macaca mulatta)25. As observed in human cells, fibroblasts isolated from younger chimpanzees allowed much more efficient MCPyV infection compared to the older animals25.
Compared to traditional FISH, HCR-DNA FISH is a simple, but highly sensitive method for detecting viral genomes24,27. Centers of MCPyV replication can be readily detected in the nuclei of infected cells as highly specific punctate foci, with little to no background signal detected in the same samples treated with a HPV specific probe (Figure 4)24. In the future, the approach could therefore be explored to detect low-abundance MCPyV DNA present in infected human skin. It could also be adapted to monitor other DNA viruses. When combined with immunofluorescent staining, the immuno-FISH provides a powerful method for simultaneously detecting viral DNA and either viral encoded protein or host proteins present in the same cells (Figure 4). For obtaining the optimum results using the protocols described here, it is best to use freshly fixed samples for immunofluorescent staining and HCR-DNA FISH analysis. For the HCR-DNA FISH analysis, the probes and amplifier should be stored in -80 °C in small aliquots to avoid repeated freezing and thawing.
Using the HDF cell culture and MCPyV infection protocol, we explored the cell growth conditions that best support MCPyV entry, transcription, and replication in HDFs. We discovered that treatment of HDFs with growth factors, such as EGF, bFGF, and stimulation of the WNT signaling pathway significantly promote MCPyV infection. Gene expression profiling reveals that, upon stimulation with these growth factors and activation of the WNT signaling pathway, several genes of the matrix metalloproteinase (MMP) family, including MMP1, 3, 7, 9, 10, 11 and 13, were robustly induced. Because these MMP enzymes are capable of degrading extracellular matrix proteins, we reason that they may stimulate MCPyV infection by disrupting the extracellular matrix of the host cells. Indeed, treating HDFs with collagenase type IV, a member of the MMP family, robustly stimulates MCPyV infection (Figure 3)24. We also screened an array of compounds, including several FDA-approved kinase inhibitors, for inhibitory effect on MCPyV infection. We discovered that trametinib, a MEK1 and MEK2 inhibitor, dramatically inhibits MCPyV infection24. Others may use or modify this infection system as a drug discovery platform for MCPyV inhibitors that reduce the MCPyV viral load in immunocompromised patients.
In summary, these protocols for achieving highly efficient MCPyV infection provide a platform to elucidate the poorly understood MCPyV infectious cycle and MCPyV-induced oncogenic mechanisms in the context of viral infection. This set of protocols could be applied in future studies to explore additional MCPyV-permissive human cell types, and the original cells of MCC, in which incidental MCPyV infection could lead to tumor development.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors would like to thank Dr. Meenhard Herlyn (Wistar Institute) and Dr. M. Celeste Simon (University of Pennsylvania) for providing reagents and technical support. We also thank the members of our laboratories for helpful discussion. This work was supported by the National Institutes of Health (NIH) grants (R01CA187718, R01CA148768 and R01CA142723), the NCI Cancer Center Support Grant (NCI P30 CA016520), and the Penn CFAR award (P30 AI 045008).
Materials
| Fetal calf serum | HyClone | SH30071.03 | |
| MEM Non-Essential Amino Acids Solution, 100X | Thermo Fisher Scientific | 11140050 | |
| GLUTAMAX I, 100X | Thermo Fisher Scientific | 35050061 | L-Glutamine |
| DPBS, no calcium, no magnesium | Thermo Fisher Scientific | 14190136 | |
| 0.05% Trypsin-EDTA | Thermo Fisher Scientific | 25300-054 | |
| DMEM/F12 medium | Thermo Fisher Scientific | 11330-032 | |
| Recombinant Human EGF Protein, CF | R&D systems | 236-EG-200 | Store at -80 degree celsius |
| CHIR99021 | Cayman Chemical | 13122 | Store at -80 degree celsius |
| CHIR99021 | Sigma | SML1046 | Store at -80 degree celsius |
| Collagenase type IV | Thermo Fisher Scientific | 17104019 | |
| Dispase II | Roche | 4942078001 | |
| Antibiotic-Antimycotic | Thermo Fisher Scientific | 15240-062 | Protect from light |
| DMEM medium | Thermo Fisher Scientific | 11965084 | |
| Alexa Fluor 594 goat anti-mouse IgG | Thermo Fisher Scientific | A11032 | Protect from light |
| Alexa Fluor 488 goat anti-rabbit IgG | Thermo Fisher Scientific | A11034 | Protect from light |
| OptiPrep Density Gradient Medium | Sigma | D1556 | Protect from light |
| Paraformaldehyde | Sigma | P6148 | |
| anti-MCPyV LT (CM2B4) | Santa Cruz | sc-136172 | Lot # B2717 |
| MCV VP1 rabbit | Rabbit polyclonal serum #10965 | https://home.ccr.cancer.gov/lco/BuckLabAntibodies.htm | |
| Hygromycin | Roche | 10843555001 | |
| Basic Fibroblast Growth Factors (bFGF), Human Recombinant | Corning | 354060 | Store at -80 degree celsius |
| Benzonase Nuclease | Sigma | E8263 | |
| Plasmid-Safe ATP-Dependent DNase | EPICENTRE | E3101K | |
| Probe hybridization buffer | Molecular technologies | ||
| Probe wash buffer | Molecular technologies | ||
| Amplification buffer | Molecular technologies | ||
| Alexa 594-labeled hairpins | Molecular technologies | B4 | Protect from light |
| Triton X-100 | Sigma | X100 | |
| Quant-iT PicoGreen dsDNA Reagent | Thermo Fisher Scientific | P7581 | |
| BamHI-HF | NEB | R3136 | |
| Buffer PB | Qiagen | 19066 | |
| blue miniprep spin column | Qiagen | 27104 | |
| 50mL Conical Centrifuge Tubes | Corning | 352070 | |
| T4 ligase | NEB | M0202T | |
| MagicMark XP | Thermo Fisher Scientific | LC5602 |
References
- Gjoerup, O., Chang, Y., Vande Woude, G., Klein, G. . Advances in Cancer Research. 106, 1-51 (2010).
- Feng, H., Shuda, M., Chang, Y., Moore, P. S. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science. 319, 1096-1100 (2008).
- Lemos, B., Nghiem, P. Merkel cell carcinoma: more deaths but still no pathway to blame. Journal of Investigative Dermatology. 127 (9), 2100-2103 (2007).
- Agelli, M., Clegg, L. X. Epidemiology of primary Merkel cell carcinoma in the United States. Journal of the American Academy of Dermatology. 49 (5), 832-841 (2003).
- Liu, W., MacDonald, M., You, J. Merkel cell polyomavirus infection and Merkel cell carcinoma. Current Opinion in Virology. 20, 20-27 (2016).
- Harrison, C. J., et al. Asymmetric Assembly of Merkel Cell Polyomavirus Large T-Antigen Origin Binding Domains at the Viral Origin. Journal of Molecular Biology. 409 (4), 529-542 (2011).
- Kwun, H. J., et al. The Minimum Replication Origin of Merkel Cell Polyomavirus Has a Unique Large T-Antigen Loading Architecture and Requires Small T-Antigen Expression for Optimal Replication. Journal of Virology. 83 (23), 12118-12128 (2009).
- Carter, J. J., et al. Identification of an overprinting gene in Merkel cell polyomavirus provides evolutionary insight into the birth of viral genes. Proceedings of the National Academy of Sciences of the United States of America. 110 (31), 12744-12749 (2013).
- Seo, G. J., Chen, C. J., Sullivan, C. S. Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression. Virology. 383 (2), 183-187 (2009).
- Theiss, J. M., et al. A Comprehensive Analysis of Replicating Merkel Cell Polyomavirus Genomes Delineates the Viral Transcription Program and Suggests a Role for mcv-miR-M1 in Episomal Persistence. PLOS Pathogens. 11 (7), 1004974 (2015).
- Schowalter, R. M., Pastrana, D. V., Buck, C. B. Glycosaminoglycans and sialylated glycans sequentially facilitate Merkel cell polyomavirus infectious entry. PLOS Pathogens. 7 (7), 1002161 (2011).
- Schowalter, R. M., Reinhold, W. C., Buck, C. B. Entry Tropism of BK and Merkel Cell Polyomaviruses in Cell Culture. PLoS One. 7 (7), 42181 (2012).
- Schowalter, R. M., Buck, C. B. The Merkel cell polyomavirus minor capsid protein. PLOS Pathogens. 9 (8), 1003558 (2013).
- Houben, R., Schrama, D., Becker, J. C. Molecular pathogenesis of Merkel cell carcinoma. Experimental Dermatology. 18 (3), 193-198 (2009).
- Chang, Y., Moore, P. S. Merkel cell carcinoma: a virus-induced human cancer. Annual Review of Pathology. 7, 123-144 (2012).
- Hodgson, N. C. Merkel cell carcinoma: Changing incidence trends. Journal of Surgical Oncology. 89 (1), 1-4 (2005).
- Tolstov, Y. L., et al. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. International Journal of Cancer. 125 (6), 1250-1256 (2009).
- Schowalter, R. M., Pastrana, D. V., Pumphrey, K. A., Moyer, A. L., Buck, C. B. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host & Microbe. 7 (6), 509-515 (2010).
- Foulongne, V., et al. Human Skin Microbiota: High Diversity of DNA Viruses Identified on the Human Skin by High Throughput Sequencing. PLoS One. 7 (6), 38499 (2012).
- Hopcraft, S. E., Damania, B. Tumour viruses and innate immunity. Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 372 (1732), (2017).
- Feng, H., et al. Cellular and viral factors regulating Merkel cell polyomavirus replication. PLoS One. 6 (7), 22468 (2011).
- Neumann, F., et al. Gene Expression and Particle Production by a Consensus Merkel Cell Polyomavirus (MCPyV) Genome. PLoS One. 6 (12), 29112 (2011).
- Tsang, S. H., Wang, X., Li, J., Buck, C. B., You, J. Host DNA damage response factors localize to merkel cell polyomavirus DNA replication sites to support efficient viral DNA replication. Journal of Virology. 88 (6), 3285-3297 (2014).
- Liu, W., et al. Identifying the Target Cells and Mechanisms of Merkel Cell Polyomavirus Infection. Cell Host & Microbe. 19 (6), 775-787 (2016).
- Liu, W., Krump, N. A., MacDonald, M., You, J. Merkel Cell Polyomavirus Infection of Animal Dermal Fibroblasts. Journal of Virology. 92 (4), (2018).
- Liu, W., et al. BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ. 21 (12), 1950-1960 (2014).
- Choi, H. M., Beck, V. A., Pierce, N. A. Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano. 8 (5), 4284-4294 (2014).
- Buck, C. B., Pastrana, D. V., Lowy, D. R., Schiller, J. T. Efficient intracellular assembly of papillomaviral vectors. Journal of Virology. 78 (2), 751-757 (2004).
- . ccr.cancer.gov Available from: https://home.ccr.cancer.gov/lco/support.htm (2018)
- . ccr.cancer.gov Available from: https://home.ccr.cancer.gov/lco/BuckLabAntibodies.htm (2018)
- . ccr.cancer.gov Available from: https://home.ccr.cancer.gov/lco/NativeMCVproduction.htm (2018)

