Isolating and Imaging Live, Intact Pacemaker Regions of Mouse Renal Pelvis by Vibratome Sectioning
Summary
The goal of this protocol is to isolate intact pacemaker regions of the mouse renal pelvis using vibratome sectioning. These sections can then be used for in situ Ca2+ imaging to elucidate Ca2+ transient properties of pacemaker cells and other interstitial cells in vibratome slices.
Abstract
The renal pelvis (RP) is a funnel-shaped, smooth muscle structure that facilitates normal urine transport from the kidney to the ureter by regular, propulsive contractions. Regular RP contractions rely on pacemaker activity, which originates from the most proximal region of the RP at the pelvis-kidney junction (PKJ). Due to the difficulty in accessing and preserving intact preparations of the PKJ, most investigations on RP pacemaking have focused on single-cell electrophysiology and Ca2+ imaging experiments. Although important revelations on RP pacemaking have emerged from such work, these experiments have several intrinsic limitations, including the inability to accurately determine cellular identity in mixed suspensions and the lack of in situ imaging of RP pacemaker activity. These factors have resulted in a limited understanding of the mechanisms that underlie normal rhythmic RP contractions. In this paper, a protocol is described to prepare intact segments of mouse PKJ using a vibratome sectioning technique. By combining this approach with mice expressing cell-specific reporters and genetically encoded Ca2+ indicators, investigators may be able to more accurately study the specific cell types and mechanisms responsible for peristaltic RP contractions that are vital for normal urine transport.
Introduction
The renal pelvis (RP) is a funnel-shaped, smooth muscle structure that transports urine from the kidney to the ureter. The RP transports urine by generating regular rhythmic contractions (peristalsis)1,2,3,4,5, which propels a bolus of urine from the kidney distally to the ureter and ultimately to the bladder, where it is stored until micturition occurs6,7. Loss of this regular activity has dire consequences, including hydronephrosis and kidney failure1,3,8; hence, there is a critical need to study the mechanisms underlying regular, rhythmic RP contractions. Peristaltic contractions originate from the most proximal region of the RP-in the pelvis-kidney junction (PKJ)9,10,11,12,13,14,15 (Figure 1A–C) and propagate distally to push urine from the papilla into the RP (Figure 1B). Electrical pacemaker activity is recorded in the PKJ as spontaneous transient depolarizations10,11,12,13,15,16,17, which are thought to arise from specialized pacemaker cells. These pacemaker cells, previously called atypical smooth muscle cells (ASMCs), are thought to generate or coordinate pacemaker activity and drive the contractions of "typical" smooth muscle cells (SMCs)9,10,11,18,19,20,21,22,23.
ASMCs are most abundant in the proximal RP, at the PKJ (Figure 1A–C), where peristaltic contractions and electrical pacemaker activity originates5,7,8,9,12,13,14,16,17,18,19,20,21,22. A recently published study by this group identified platelet-derived growth factor receptor-alpha (PDGFRα), in combination with smooth muscle myosin heavy chain (smMHC), as a unique biomarker for these interstitial cells (ICs)24, a finding that has been corroborated by other groups25. Based on their morphology and protein expression pattern, these cells were called PDGFRα+ IC type 1 (PIC1)24,26. PIC1s reside in the muscle layer of the PKJ where they display high-frequency, short-duration Ca2+ transients, thought to underlie the generation of pacemaker potentials24. However, other cell types exist in the PKJ, including non-smMHC-expressing PDGFRα+ ICs (PIC2s) in the adventitial layer. Previous reports have suggested that non-smMHC ICs may participate in the regulation of pacemaker activity19. However, further study of non-smMHC ICs is hindered by poor distinction during Ca2+ imaging studies. Typically, heterogeneous cell types within the RP preparations are indiscriminately loaded with Ca2+-sensitive dyes (e.g., Fluo-4). To overcome these challenges and to study a variety of cell types in the RP, genetically encoded Ca2+ indicators (GECIs) can be utilized to selectively express Ca2+-sensitive fluorophores in cell types of interest.
The majority of studies elucidating Ca2+ transient properties in PIC1s/ASMCs were achieved by imaging flat-sheet RP tissue preparations19,21,27. As PIC1s are the only cell type in the PKJ to express smMHC, conditional expression of the GECI, GCaMP, in smMHC+ cells is appropriate to study PIC1s in this configuration. However, as PIC1s and PIC2s both express PDGFRα, conditional expression of GCaMP variants in PDGFRα+ cells prohibit cell distinction in flat-sheet preparations. To circumvent this issue, a vibratome sectioning approach was used to distinguish PIC1s and PIC2s across the PKJ tissue wall24. To reveal these discrete cellular populations, the RP was sectioned coronally, making it possible to identify PIC2s in the adventitia and PIC1s in the muscle wall based on known immunohistochemical labeling and GECI expression patterns. As a result of this novel PKJ imaging approach, PIC1s and PIC2s were found to display distinct Ca2+ signaling properties24. Furthermore, by isolating the most proximal sections of the PKJ region (Figure 2), the pacemaker region of the RP was preserved in a way that had not been accomplished previously. Here, a protocol is described to show how to isolate PKJ preparations from the mouse kidney using vibratome sectioning, how to set up these preparations for Ca2+ imaging experiments, and how to distinguish the different cell types across the PKJ wall.
Protocol
All mice used and the protocols described in this study were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and the Institutional Animal Use and Care Committee at the University of Nevada, Reno, NV. Experiments and animal use also conformed to the principles and regulations as described by Grundy28.
1. Generate PDGFRα-GCaMP6f and SMCGCaMP3 mice
- Cross GCaMP6flox/+ mice (B6; 129S-Gt(ROSA)26Sortm95.1(CAG-GCaMP6f)Hze/J) with PDGFRαCre mice (C57BL/6-Tg(Pdgfra-cre)1Clc/J) to generate PDGFRα-GCaMP6f mice.
- Cross GCaMP3lox/+ mice (B6.129S-Gt(ROSA)26Sortm38(CAG-GCaMP3)Hze/J) with male smMHCCreERT2 mice (B6.FVB-Tg(Myh11-cre/ERT2)1Soff/J) to generate SMC-GCaMP3 mice.
NOTE: Only male mice of the cross (GCaMP3lox/+ mice and smMHCCreERT2 mice) can be used as Cre expression is driven from the Y chromosome. smMHCCreERT2 mice can also be crossed with GCaMP6flox/+ mice for improved Ca2+ signal-to-noise ratio and faster Ca2+ signal temporal properties.
2. Prepare and inject transgenic mice with tamoxifen to induce conditional expression of GCaMP
- Activate inducible Cre recombinase for cell-specific GCaMP expression in specific cell types, as previously described29.
3. Prepare solutions
- Prepare 1 L of Krebs-Ringer bicarbonate (KRB) solution containing 120.35 mM NaCl, 5.9 mM KCl, 15.5 mM NaHCO3, 1.2 mM Na2HPO4,1.2 mM MgCl2, 11.5 mM glucose, and 2.5 mM CaCl2. On the day of use, maintain the KRB solution on ice.
NOTE: KRB can be stored at 4 °C for up to one week and should be pre-bubbled with a mix of 97% O2 and 3% CO2 for at least 10 min prior to use.
4. Prepare silicon elastomer-coated dissection and microscope imaging dishes
- Mix silicon elastomer components according to the manufacturer's instructions. Fill a 35 mm x 10 mm Petri dish and a 60 mm x 15 mm Petri dish approximately one-fourth full of liquid silicon elastomer for imaging experiments and dissection, respectively. Polymerize the silicon elastomer at 37 °C for 1 day prior to use.
NOTE: To enhance the contrast of thin kidney sections, apply a small, black circle of paper to the base of the imaging Petri dish before filling with silicon elastomer.
5. Kidney dissection
- Anaesthetize mice by inhalation of 3-4% isoflurane in a ventilated hood. Confirm the induction of deep anesthesia by loss of toe and/or tail pinch reflex, and then euthanize the mice by cervical dislocation.
- Apply 70% ethanol to the chest to dampen the fur. Using external dissection scissors, open the abdominal cavity via a longitudinal incision, with scissor blades angled away from the animal to prevent damage to the internal organs.
- Using internal tissue forceps and internal dissection scissors, pinch the intestines and lift them away from the abdominal wall. Whilst lifting the intestines, cut the underside of the intestines free from the body at the proximal duodenum and distal colon to gain access to the retroperitoneal space containing the kidneys.
- Once the kidneys are exposed, extract them individually. Gently pinch and lift the distal end of the ureter (~4mm away from the kidney) with tissue forceps. Using the dissection scissors, cut underneath the pinched ureter towards the kidney. Continue to cut underneath the kidney until it has become liberated from the surrounding connective tissue.
NOTE: To maximize tissue integrity and cutting consistency during vibratome sectioning, the kidney must be as intact as possible. To ensure this, avoid pinching or cutting of the kidney with forceps and dissection scissors. - Place the kidney with attached ureter in ice-cold KRB solution. Repeat steps 5.4 and 5.5 with the contralateral kidney. Maintain the kidneys in KRB solution on ice.
NOTE: Immediately proceed to the next section of the protocol to preserve PKJ tissue viability. Because of its anatomical location deep in the kidney parenchyma, the PKJ is deprived of contact with KRB solution.
6. Prepare the kidney for vibratome sectioning
- Transfer the kidney to a silicon elastomer-coated dissection dish (60 mm x 15 mm), and fill it with ice-cold KRB solution until the kidney is completely submerged.
- Under a dissecting microscope, anchor the kidney to the base of the dish by inserting minutien pins into the proximal ureter and through the thin anterior renal capsule or surrounding adipose tissue.
NOTE: Take care not to puncture the kidney parenchyma tissue. - Use fine spring scissors and internal forceps to remove adipose tissue from the base of the kidney to expose the distal RP and proximal ureter.
- Remove the proximal ureter and a portion of the distal RP from the base of RP using fine spring scissors.
NOTE: Take care not to cut the surrounding kidney parenchyma. The black dashed line in Figure 1A indicates the approximate position of this cut. This cut creates a flat base on the kidney for more uniform tissue sectioning. When dissecting the kidney, one must be aware of the anatomical position of the PKJ region. Figure 1B shows that the intact kidney can be cut along a sagittal plane to expose the medulla, papilla (distal medulla where collecting ducts converge) and proximal and distal RP. If the papilla were to be exposed completely, as in Figure 1C, the PKJ and proximal RP (prox RP) can be visualized. However, this should not be done for the vibratome technique; this description is to orientate the reader to the PKJ location generally, emphasized in the anatomical difference shown in the transmitted light images of the PKJ region and mid-RP region in Figure 1D,E. - Pierce the outer renal capsule with fine-tip forceps, angling the tips away from the kidney body. Using forceps with each hand, pinch the loose ends of the capsule and peel them apart. Continue to peel back the remaining renal capsule membrane until it is removed entirely.
NOTE: The renal capsule is a tough, fibrous layer that surrounds the kidney. It must be removed prior to vibratome sectioning for optimal cutting.
7. Prepare and calibrate the vibratome instrument
- Insert a razor blade into the blade holder of the vibratome instrument, and adjust the blade clearance angle to ~18°.
NOTE: As an optional step for higher quality sectioning, a calibration block (provided with some vibratome instruments) should be used to adjust the blade position for each new blade used according to the manufacturer's instructions. This will ensure optimum positioning of the blade and minimize vertical vibration. - Adjust the blade advancement speed to 0.2 mm/s, the horizontal vibration/sheering of the blade to an amplitude of 2.00 mm, and the Z-step size of the blade to ~100-150 µm. Ensure that kidney section thickness does not exceed 150 µm as this will negatively impact Ca2+ imaging experiments (because the PKJ wall will often roll and fold on itself if it is too thick).
NOTE: The user should fine-tune these cutting parameters because settings will vary between individual kidney preparations and vibratome instruments. Fine-tuning should take place during sectioning when the user can visually inspect sections under a microscope, as described in step 7.2. - Install an ice bath and buffer tray onto the vibratome instrument. Fill the ice bath with crushed ice, and fill the inner stage area with ice-cold KRB solution (fill to approximately half-full). During vibratome sectioning, monitor and replace the crushed ice that has melted.
8. Vibratome sectioning the kidney
- Use blunt-ended forceps to gently grasp and remove the prepared kidney from ice-cold KRB solution. Immediately place the kidney on absorbent paper for ~2-4 s to remove excess external moisture. Gently roll the kidney across the absorbent paper to ensure that all sides of the parenchyma have dried so that there is optimal adhesion of the kidney to the vibratome stage.
NOTE: As the RP is located inside the kidney and therefore protected by the outer parenchyma, this short drying period would not be detrimental to tissue integrity. - Immediately apply a thin layer of cyanoacrylate glue (~1 cm2) to the base of the vibratome specimen plate, and use blunt-ended forceps to place the kidney, ureter side down, on the area covered in glue. Gently apply downward pressure to the top of the kidney with the flat edge of the forceps for approximately 10-20 s to dry the glue.
NOTE: To stabilize the kidney during this procedure, use an additional pair of forceps to keep the kidney upright as the glue dries. It is critical that the kidney adheres to the specimen plate in an upright position so that sections are cut straight. To ensure that the kidney has successfully adhered to the specimen plate, gently push the side of the kidney. If the kidney has successfully adhered to the plate, the base of the kidney should stay secured to the plate. - Firmly secure the specimen plate to the bottom of the buffer tray. Adjust the level of KRB solution so that the top of the kidney is fully immersed. During sectioning steps, as the vibratome blade moves deeper into the buffer tray, remove KRB solution so that the blade holder does not become immersed in solution.
NOTE: If the glue has not completely dried before proceeding with this step, the glue will often move up from the base and on to the kidney parenchyma. This excess glue will make sectioning more variable. If this does happen, remove the kidney from the plate and proceed with a fresh kidney preparation. - For automatic vibratome sectioning, select the start and end positions of the vibratome blade-cutting cycle. Verify these positions are ~0.5-1 cm clear of the kidney to ensure that with each blade advancement, the entire kidney plane is sectioned.
- Prepare a multi-welled plate (24- or 48-well) by filling wells with KRB solution and place the plate on ice.
NOTE: When generated, individual sections should be placed into separate wells to keep track of section depth. - Start the automatic cutting process. During the initial pass of the blade, ensure that the blade makes contact with the very top of kidney. If contact is not made, adjust the starting Z-position of the blade.
- Using forceps, collect sections that are liberated from the kidney. Immediately transfer the sections to individual wells, and note the Z-depth of the sections to gauge the approximate PKJ location within kidney sections.
NOTE: Depending on the cut parameters, some sections may not be cut free from the kidney block. If this occurs, carefully use fine spring scissors to cut sections from the kidney block. Users are also encouraged to actively visualize sections under a light microscope whilst free-floating in individual wells to ensure optimal cut settings and tissue location. Sections containing the PKJ will typically be derived ~ 1000-1500 µm from the top of the kidney. - Continue the sectioning protocol until the PKJ regions become more apparent. Refer to the representative results section for description of the PKJ regions as sectioning proceeds.
- At this point, optimize the sectioning parameters to ensure that sections are liberated from the kidney block uniformly and are intact. Additionally, make sure that the PKJ regions are continuous and unbroken because broken PKJ walls will not allow adequate imaging of cells within the wall due to collapse. If walls become broken, decrease the sectioning speed and increase the section thickness, and continue to observe sections under a light microscope to fine-tune the cutting parameters.
- Store sections at 4°C in KRB solution until experimentation begins.
9. Kidney slice Ca2+ image acquisition
- Transfer an individual kidney slice to a silicon elastomer-coated imaging dish (35 mm x 10 mm), and immediately fill the dish with fresh, ice-cold KRB solution.
- Insert minutien pins around the periphery of a kidney slice to secure the section to the base of the imaging dish.
NOTE: This step is critical to prevent the section from moving around when physiological solutions are perfused over the slice during imaging. - Place the imaging dish on the stage of an upright spinning-disk confocal microscope and immediately start perfusing with KRB solution.
NOTE: In this protocol, an upright microscope equipped with a high-speed Nipkow spinning disk confocal scanner unit was used. - Maintain the perfusion rate at 3 mL/min and KRB solution temperature at 36 ± 1 °C. Prior to imaging, allow the slice to equilibrate for 1 h.
- Select the appropriate dichroic imaging cube and lasers. Acquire images with an electron-multiplying charge-coupled device (EMCCD) or scientific complementary metal-oxide-semiconductor camera.
NOTE: The confocal system in this protocol is equipped with a 488 nm laser to excite GCaMP6f or GCaMP3. Images were acquired using a 512 pixel x 512 pixel EMCCD camera. - Use a low magnification, water-immersion objective lens (4x or 10x) to locate the kidney slice. Center the imaging field on areas of the slice where the PKJ is present. Identify landmarks, as depicted in Figure 2D, to locate the PKJ (i.e., semicircles of muscular tissues suspended between parenchymal tissue).
- Once the PKJ is located, use a higher magnification water-immersion objective lens (20x, 40x, or 60x) to magnify the area of interest.
NOTE: In this protocol, the 20x objective numerical aperture (NA) was 1.0, the 40x objective NA was 0.8, and the 60x objective NA was 1.0. - Distinguish different cells of interest in the PKJ wall using transgenic mice expressing GCaMP6f or GCaMP3 in PDGFRα+ cells or SMCs, respectively. Observe the different types of Ca2+ transient durations in PDGFRα+ cells in the PKJ wall in PDGFRα+ GCaMP6f+ kidney slices (Figure 3C, see representative results for description) and in GCaMP3+ SMCs in SMC GCaMP3+ kidney slices (Figure 3D, see representative results for description).
- Once cells of interest in PKJ wall have been identified, adjust the laser intensity to yield a good signal-to-noise ratio. Record images at a temporal sampling frequency between 16 Hz and 32 Hz using appropriate acquisition software.
NOTE: Depending on laser intensity used during imaging, it is recommended to limit the number of recordings owing to the bleaching effects on Ca2+ fluorophores. The user should choose recording length based upon the specific experimental objectives.
10. Ca2+ imaging analysis
- Perform Ca2+ imaging analysis of different cell types in PKJ pacemaker regions by spatiotemporal mapping as previously described for other intact pacemaker preparations in the gastrointestinal tract29,30.
Representative Results
In situ Ca2+ imaging of the PKJ can reveal important cellular activity of RP pacemaker cells. By using mice that express genetically encoded Ca2+ indicators (such as GCaMP), driven by cell-specific promotors, information about RP pacemaking can be obtained with accuracy and detail that is not possible from Ca2+ imaging experiments from flat-sheet RP preparations. The beginning of the PKJ is distinguished by the sudden appearance of semi-circles of muscle suspended between kidney parenchymal tissue (Figure 2C; proximal PKJ enclosed in dashed box). During subsequent rounds of sectioning, the inner medulla becomes distinguishable from the surrounding cortical tissue. Under a light microscope, the inner medulla appears striated in regions, lighter in color compared to cortical tissue and discontinuous on its long axis with the rest of the kidney (Figure 2B,D). At this point, more PKJ regions will start to appear. Examples of this are shown in Figure 2D (dashed rectangles, H and G) where 3 semi-circles of muscle are suspended by parenchymal tissue. These muscle bands will be closely apposed to the inner papilla and will typically neighbor a renal arteriole (Figure 2D, dashed rectangles; Figure 2F–H, black arrowheads). As more distal sections are derived, these bands of muscle will integrate to form a more complete, unified structure, indicating the end of the PKJ region (Figure 2E).
Figure 3A,B shows a PKJ section at low power (4-10x) from a mouse expressing GCaMP in PDGFRα+ cells (GCaMP6f expressed by inducible Cre-recombinase driven by Pdgfra). Using landmarks such as the renal arteriole (Figure 3A; asterisk), experimenters should be able to readily distinguish the thin PKJ wall suspended between parenchymal tissue (Figure 3B; asterisks). The expression of GCaMP6f in this specific transgenic tissue is spread across the entire width of the PKJ, across both the muscle and adventitial layers (Figure 3C).
In PDGFRα+ GCaMP6f+ kidney slices, a network of cells that typically extends over the width of the PKJ wall will be fluorescent (Figure 3C) and display oscillating Ca2+ transients of various durations and frequencies. PDGFRα+ cells in the PKJ wall display two different types of Ca2+ transient durations. In the adventitial layer (orientated closer to cortex), PDGFRα+ cells present as a network of cells and their processes are defined. Adventitial PDGFRα+ cells exhibit low-frequency (4 ± 2.7 Hz) and long-duration (1 ± 0.67 s) Ca2+ transients. The second layer of PDGFRα+ cells, present in the muscle layer (orientated closer to the medulla), exhibit similar Ca2+ transient frequencies and durations as SMC GCaMP3+ cells (described below) as they are the same cell type.
In SMC GCaMP3+ kidney slices, a layer of GCaMP3+ cells is present in the muscle layer (Figure 3D). There will be no fluorescent signal in the adventitial layer (Figure 3D; asterisk). GCaMP3+ SMCs in the muscular layer typically exhibit high-frequency (10 ± 4 Hz) and short-duration (632 ± 74 s) Ca2+ transients. PDGFRα+ cells located in the PKJ adventitia elicit long-duration, low-frequency Ca2+ transients (Figure 3E, Video 1). However, Ca2+ imaging experiments from tissue expressing GCaMP3 driven by the Myh11 promotor is restricted to the muscular aspect of the PKJ (Figure 3D). Compared to PDGFRα+ cells in the adventitia, SMCs fired shorter duration Ca2+ transients more frequently (Figure 3F, Video 2).
In addition to understanding signaling properties in PKJ PDGFRα+ ICs, the application of this technique to study other cell types in the vibratome-sectioned kidney has been demonstrated in this paper. Upon close examination of the renal medulla (in mice expressing GCaMP6f in PDGFRα+ cells), an array of fluorescent Ca2+ signals within and surrounding collecting ducts (Video 3) was observed. Medullary PDGFRα+ cells fired spontaneous Ca2+ transients of variable frequency and duration. These Ca2+ imaging studies of kidney vibratome sections could also be expanded to studying renal arterioles (~50-80 mm diameter) that often neighbor PKJ muscle segments (Figure 2F,G; white arrows). Ca2+ imaging of renal arterioles (from tissue expressing GCaMP in smooth muscle cells) demonstrates oscillating Ca2+ transients in SMCs (Video 4).
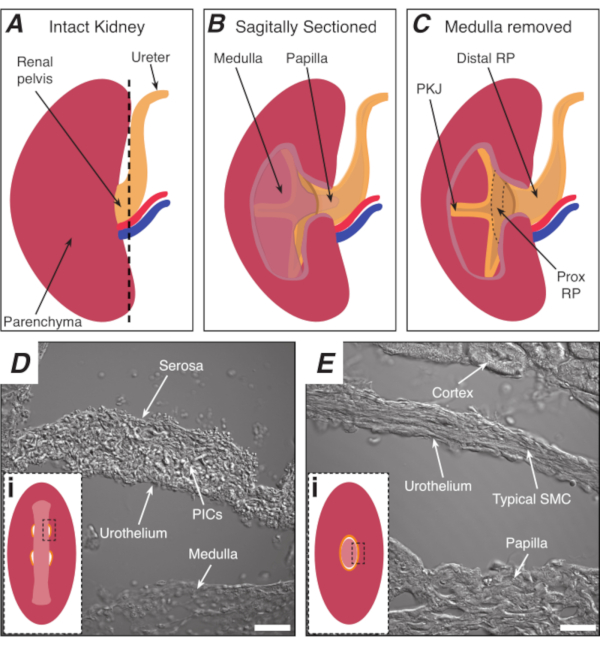
Figure 1: Basic kidney anatomy and location of PKJ pacemaker region. (A) Diagram of the intact kidney showing the orientation of the RP and ureter. The renal artery and renal vein are displayed in red and blue, respectively. (B) The intact kidney can be cut along a sagittal plane to expose the inner aspect of the kidney, including the medulla, papilla (distal medulla where collecting ducts converge), and proximal and distal RP. (C) The medulla and papilla can be excised to completely expose the PKJ and prox RP. (D and E) represent transmitted light images from the PKJ pacemaker region and distal RP, respectively. Sequential sectioning towards the distal end of the pelvis results in the semicircles of muscle in the PKJ region (Di) combining into one, thicker muscular ring (Ei) that encapsulates the entire papilla. Black, dashed rectangles in Di and Ei show approximate areas in coronal kidney sections where transmitted light images were acquired. Orientation of images D and E are 90° anti-clockwise to respective insets (Di and Ei). Scale bars in D and E = 20 µm. Abbreviations: RP = renal pelvis; prox RP = proximal renal pelvis; PKJ = pelvic-kidney junction; PICs = platelet-derived growth factor receptor-alpha-positive interstitial cells; SMC = smooth muscle cell. Please click here to view a larger version of this figure.
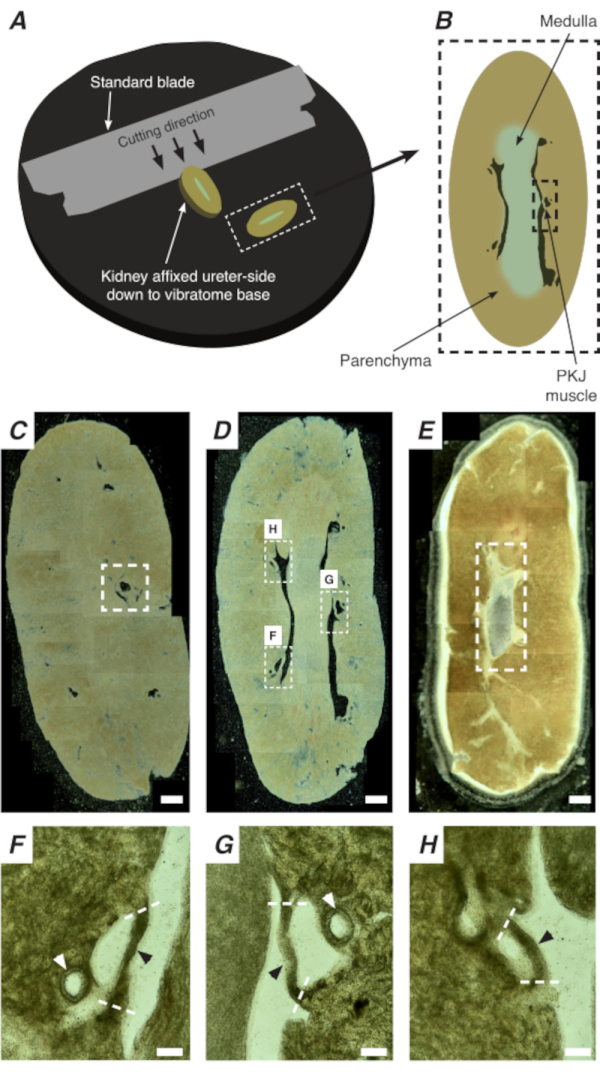
Figure 2: Vibratome sectioning of whole kidneys to generate thin sections. (A) Kidneys are mounted ureter side down to the base of the vibratome instrument, and a standard blade (attached to the vibratome head) is used to cut sequential sections from the proximal to distal end of the kidney. (B) Diagrammatic representation of a thin section cut from the whole kidney with annotated landmarks. PKJ muscle segments (black dashed rectangle) are often found suspended between parenchymal tissue. (C) Light microscopic image of a proximal kidney section. The appearance of muscle bands suspended between parenchymal tissue indicates the beginning of the proximal PKJ projections (indicated inside white dashed rectangle). (D) Light microscopic image representing the optimal region where multiple (2-3) PKJ segments can be found (areas within white dashed rectangles). Thin PKJ muscle strips are suspended between the kidney parenchyma and align closely with renal arterioles and medulla. (E) Light microscopic image of a distal kidney section. Individual muscle segments have merged to form a single, continuous muscle band (white dashed rectangle) that surrounds the inner papilla (not present in this image). Scale bars C–E = 500 µm. F–H Zoomed (20x) regions from panel D indicate the location of the PKJ (black arrowheads), renal arterioles (white arrowheads), and cut sites for isolating the PKJ (dashed white lines). Scale bars F–H = 100 µm. Abbreviation: PKJ = pelvic-kidney junction. Please click here to view a larger version of this figure.
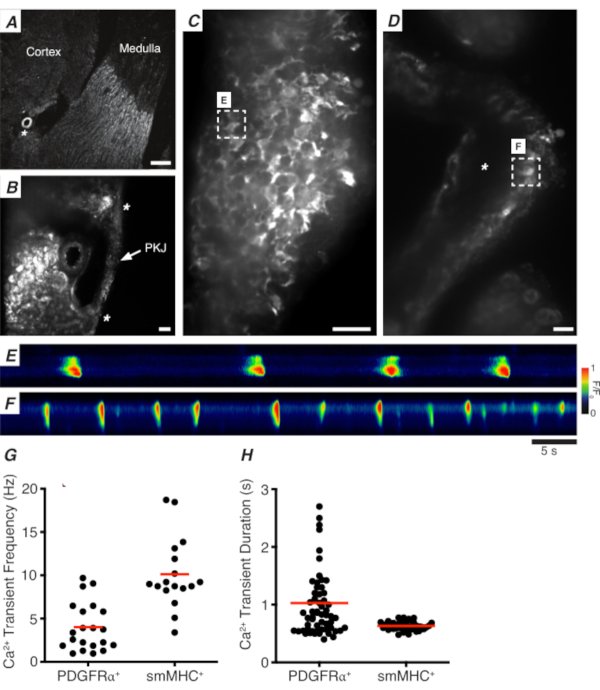
Figure 3: Ca2+ imaging of vibratome sections. (A) Representative low-power image (4x) of a vibratome section denoting location of the renal arteriole (asterisk). Scale bar = 200 µm. (B) Zoomed (20x) representative image of the PKJ (labeled) suspended between kidney parenchymal tissue denoting locations of the PKJ muscle (white arrowhead), renal arteriole (asterisk). Scale bar = 50 µm. (C) High-power (40x) image of the PKJ expressing GCaMP in PDGFRα+ cells. Scale bar = 20 µm. (D) High-power (20x) image of the PKJ expressing GCaMP in smooth muscle cells. Scale bar = 20 µm. (E) Spatiotemporal map of Ca2+ transients sampled from a GCaMP+ PDGFRα+ cell indicated in panel C. Look up table coded for F/F0. (F) Spatiotemporal map of Ca2+ transients sampled from a GCaMP+ PDGFRα+ cell indicated in panel D. Look up table coded for F/F0. (G) Representative data for Ca2+ transient frequency (Hz) for GCaMP+ PDGFRα+ cells and GCaMP+ smMHC cells. (H) Representative data for Ca2+ transient duration (s) for GCaMP+ PDGFRα+ cells and GCaMP+ smMHC cells. Abbreviations: PKJ = pelvic-kidney junction; PDGFRα+ = platelet-derived growth factor receptor-alpha-positive; smMHC = smooth muscle myosin heavy chain. Please click here to view a larger version of this figure.
Video 1: Spontaneous Ca2+ transients in GCaMP+ PDGFRα+ cells in PKJ vibratome sections. Abbreviations: PKJ = pelvic-kidney junction; PDGFRα+ = platelet-derived growth factor receptor-alpha-positive. Please click here to view this video. (Right-click to download.)
Video 2: Spontaneous Ca2+ transients in GCaMP+ smooth muscle cells in pelvic-kidney junction vibratome sections. Please click here to view this video. (Right-click to download.)
Video 3: Spontaneous Ca2+ transients in GCaMP+ PDGFRα+ cells in renal medullary vibratome sections. Abbreviation: PDGFRα+ = platelet-derived growth factor receptor-alpha-positive. Please click here to view this video. (Right-click to download.)
Video 4: Low-amplitude Ca2+ transient activity in the vibratome sections of the renal arteriole. Please click here to view this video. (Right-click to download.)
Discussion
The RP consists of a heterogeneous population of cells with differential cell densities observed in various RP regions. PIC1s (previously referred to as ASMCs) are most abundant in the PKJ, where pacemaker activity originates24. The protocol, described here, allows investigators to isolate the pacemaker region from the rest of the mouse kidney. By cutting sections of PKJ using a vibratome, the pacemaker regions of the RP (identified as muscle bands attached to parenchyma) are kept intact, thus affording the use of in situ imaging to accurately study RP pacemaker cells when combined with cell-specific fluorescence reporters.
While this approach can provide new insights into RP pacemaking, there are some considerations that experimenters should be familiar with to improve imaging outcomes and sectioning efficiency. For an untrained user, this method of PKJ isolation and imaging is easier to learn than typical sharp dissection of flat-sheet RP preparations. Sharp dissection of RP from whole kidneys requires weeks of consistent practice to successfully isolate viable tissues for physiology experiments. As this vibratome sectioning protocol requires little sharp dissection knowledge, it is accessible to users who do not have experience dissecting other smooth muscle structures.
However, there are some critical points to note for this protocol. Successfully adhering kidneys to the vibratome specimen plate requires dexterity and patience. If the kidney is orientated incorrectly and leans to one side, oblique rather than straight sections will be cut. Due to the delicate nature of the PKJ, the oblique angle can often destroy the muscle bands of the pacemaker region. Furthermore, imaging of oblique sections results in poor imaging acquisition as cell networks are not typically in the same focal plane. The procedure is also time-consuming, with sectioning of a single kidney often taking up to an hour to complete, during which time the setup requires monitoring.
While the movement of the vibratome can be sped up, if the speed is increased too much (>20% than what is recommended in the protocol), the blade will shred rather than cleanly cut the kidney, resulting in loss of delicate PKJ structures. Similarly, a cutting speed that is too low can cause the section to become jagged. Optimization of cutting speed and blade amplitude is essential. Care must also be taken in handling vibratome sections. Due to their delicate nature, PKJ muscles are easily disrupted during handling and can tear. A well-trained user will be able to harvest approximately 1-2 PKJ regions per 4 kidney slices that are suitable for Ca2+ imaging experiments. Typically, PKJ sections that do not meet Ca2+ imaging criteria have: 1) poor GCaMP expression, 2) a contorted PKJ wall, or 3) a broken PKJ wall. For data analysis, approximately 3-4 cells per field of view (FOV) could be sampled.
Whilst there are many cells in the FOV of PDGFRα-GCaMP6f and SMC-GCaMP3 PKJ sections, small tissue movements often exclude cells from analysis. This can usually be resolved by applying a stabilization protocol to images. Under conditions where preparations do not move, at least 3-5 cells can be sampled from PDGFRα-GCaMP6f sections and 5-6 cells from SMC-GCaMP3 sections. Typically, the time taken from animal sacrifice (optimum age for mice is 8-16 weeks) to performing Ca2+ imaging experiments is 2-3 h, which is adequate to ensure tissue integrity, if tissues are incubated in ice-cold solutions throughout the procedure when required. In summary, a vibratome cutting protocol has been described here to generate intact preparations of RP PKJ regions from the mouse kidney. This technique allows the preservation of RP pacemaker regions for in situ Ca2+ imaging studies to investigate RP pacemaker mechanisms.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This project was funded by R01 DK124509 from NIDDK.
Materials
| 24-well culture plate | ThermoFisher Scientific | 142485 | To store kidney slices in |
| 35 mm x 10 mm Petri dishes | Sigma Aldrich | CLS430165 | Kidney slice calcium imaging dish |
| 48-well culture plate | ThermoFisher Scientific | 152640 | To store kidney slices in |
| 60 mm x 15 mm Petri dish | Sigma Aldrich | P5481 | Kidney sharp dissection dish |
| Absorbent paper | Fisher Scientific | 06-666A | To dry the kidney before applying glue |
| B6;129S-Gt(ROSA)26Sor/J | The Jackson Laboratory | 13148 | GCaMP3 Mice |
| B6;129S-Gt(ROSA)26Sor/J | The Jackson Laboratory | 24105 | GCaMP6f Mice |
| B6.FVB-Tg(Myh11-cre/ERT2)1Soff/J | The Jackson Laboratory | 19079 | smMHC-CRE Mice |
| C57BL/6-Tg(Pdgfra-cre)1Clc/J | The Jackson Laboratory | 13148 | PDGFRa-CRE Mice |
| Cyanoacrylate glue | Amazon | B001PILFVY | For adhering the kidney to the specimen plate |
| Ethanol | Phamco-Aaper | SDA 2B-6 | For dissection |
| Extra-Fine Bonn Scissors | Fine Science Tools | 14083-08 | Used for internal dissecting scissors |
| Fine scissors | Fine Science Tools | 14060-09 | Used for external dissecting scissors |
| Fine-tip forceps | Fine Science Tools | 11254-20 | Used for fine dissection of kidney |
| Gillette Silver Blue double-edge blades | Amazon | B009XHQGYO | For insertion into blade holder of vibratome |
| ImageJ | NIH | For calcium imaging analysis | |
| Isoflurane | Baxter | NDC 1001936060 | For anesthesia |
| Minutien pins | Fisher Scientific | NC9677548 | Pins were cut in half to reduce their length |
| Silicon elastomer | Fisher Scientific | NC9285739 | Sylgard 184 |
| Student Adson Forceps | Fine Science Tools | 91106-12 | For gently holding and moving the kidney |
| Student Dumont Forceps | Fine Science Tools | 91150-20 | Used for internal dissecting forceps |
| Vannas spring scissors | Fine Science Tools | 15000-03 | For sharp dissection and cleanup of isolated kidney |
| Vibrocheck | Leica | 14048142075 | Optional component for calibrating blade movement during cutting |
| VT1200 S Vibrating Blade Microtome | Leica | 14912000001 | Configuration 1 is used in our protocol |
References
- Constantinou, C. E., Djurhuus, J. C. Pyeloureteral dynamics in the intact and chronically obstructed multicalyceal kidney. The American Journal of Physiology. 241 (5), 398-411 (1981).
- Constantinou, C. E., Yamaguchi, O. Multiple-coupled pacemaker system in renal pelvis of the unicalyceal kidney. TheAmerican Journal of Physiology. 241 (5), 412-418 (1981).
- Constantinou, C. E., Hrynczuk, J. R. Urodynamics of the upper urinary tract. Investigative Urology. 14 (3), 233-240 (1976).
- Schmidt-Nielsen, B., Schmidt-Nielsen, B. On the function of the mammalian renal papilla and the peristalsis of the surrounding pelvis. Acta Physiologica. 202 (3), 379-385 (2011).
- Dwyer, T. M., Schmidt-Nielsen, B. The renal pelvis: machinery that concentrates urine in the papilla. Physiology. 18 (1), 1-6 (2003).
- Hill, W. G. Control of urinary drainage and voiding. Clinical Journal of the American Society of Nephrology. 10 (3), 480-492 (2015).
- Brading, A. F. The physiology of the mammalian urinary outflow tract. Experimental Physiology. 84 (1), 215-221 (1999).
- Dixon, J. S., Gosling, J. A. The musculature of the human renal calices, pelvis and upper ureter. Journal of Anatomy. 135, 129-137 (1982).
- Lang, R. J., et al. Pyeloureteric peristalsis: role of atypical smooth muscle cells and interstitial cells of Cajal-like cells as pacemakers. The Journal of Physiology. 576, 695-705 (2006).
- Lang, R. J., Takano, H., Davidson, M. E., Suzuki, H., Klemm, M. F. Characterization of the spontaneous electrical and contractile activity of smooth muscle cells in the rat upper urinary tract. Journal of Urology. 166 (1), 329-334 (2001).
- Lang, R. J., Exintaris, B., Teele, M. E., Harvey, J., Klemm, M. F. Electrical basis of peristalsis in the mammalian upper urinary tract. Clinical and Experimental Pharmacology & Physiology. 25 (5), 310-321 (1998).
- Morita, T., Ishizuka, G., Tsuchida, S. Initiation and propagation of stimulus from the renal pelvic pacemaker in pig kidney. Investigative Urology. 19 (3), 157-160 (1981).
- Tsuchida, S., Morita, T., Harada, T., Kimura, Y. Initiation and propagation of canine renal pelvic peristalsis. Urologia Internationalis. 36 (5), 307-314 (1981).
- Yamaguchi, O. A., Constantinou, C. E. Renal calyceal and pelvic contraction rhythms. American Journal of Physiology – Regulatory Integrative and Comparative Physiology. 257 (4), 788-795 (1989).
- Klemm, M. F., Exintaris, B., Lang, R. J. Identification of the cells underlying pacemaker activity in the guinea-pig upper urinary tract. Journal of Physiology. 519 (3), 867-884 (1999).
- Lang, R. J., et al. Spontaneous electrical and Ca2+ signals in the mouse renal pelvis that drive pyeloureteric peristalsis. Clinical and Experimental Pharmacology & Physiology. 37 (4), 509-515 (2010).
- Lutzeyer, W. Pacemaker process of ureteral peristalsis in multicalyceal kidneys. Urologia Internationalis. 37 (4), 240-246 (1982).
- Lang, R. J., Hashitani, H. Pacemaker mechanisms driving pyeloureteric peristalsis: modulatory role of interstitial cells. Advances in Experimental Medicine and Biology. 1124, 77-101 (2019).
- Hashitani, H., et al. Interstitial cell modulation of pyeloureteric peristalsis in the mouse renal pelvis examined using FIBSEM tomography and calcium indicators. Pflugers Archiv: European Journal of Physiology. 469 (5-6), 797-813 (2017).
- Lang, R. J., Hashitani, H., Tonta, M. A., Suzuki, H., Parkington, H. C. Role of Ca2+ entry and Ca2+ stores in atypical smooth muscle cell autorhythmicity in the mouse renal pelvis. British Journal of Pharmacology. 152 (8), 1248-1259 (2007).
- Lang, R. J., Hashitani, H., Tonta, M. A., Parkington, H. C., Suzuki, H. Spontaneous electrical and Ca2+ signals in typical and atypical smooth muscle cells and interstitial cell of Cajal-like cells of mouse renal pelvis. The Journal of Physiology. 583, 1049-1068 (2007).
- Hashitani, H., Lang, R. J., Mitsui, R., Mabuchi, Y., Suzuki, H. Distinct effects of CGRP on typical and atypical smooth muscle cells involved in generating spontaneous contractions in the mouse renal pelvis. British Journal of Pharmacology. 158 (8), 2030-2045 (2009).
- Iqbal, J., et al. Potassium and ANO1/ TMEM16A chloride channel profiles distinguish atypical and typical smooth muscle cells from interstitial cells in the mouse renal pelvis. British Journal of Pharmacology. 165 (7), 2389-2408 (2012).
- Grainger, N., et al. Identification and classification of interstitial cells in the mouse renal pelvis. Journal of Physiology. 598 (15), 3283-3307 (2020).
- Hashitani, H., Mitsui, R., Lang, R. Functional heterogeneity of PDGFRα (+) cells in spontaneously active urogenital tissues. Neurourology and Urodynamics. 39 (6), 1667-1678 (2020).
- Hashitani, H., Lang, R. J. ATYPICAL or INTERSTITIAL, take your PIC. Journal of Physiology. 598 (15), 3061-3062 (2020).
- Lang, R. J., Hashitani, H., Tonta, M. A., Suzuki, H., Parkington, H. C. Role of Ca2+ entry and Ca2+ stores in atypical smooth muscle cell autorhythmicity in the mouse renal pelvis. British Journal of Pharmacology. 152 (8), 1248-1259 (2007).
- Grundy, D. Principles and standards for reporting animal experiments in The Journal of Physiology and Experimental Physiology. Experimental Physiology. 100 (7), 755-758 (2015).
- Drumm, B. T., Hennig, G. W., Baker, S. A., Sanders, K. M. Applications of spatio-temporal mapping and particle analysis techniques to quantify intracellular Ca 2+ signaling in situ. Journal of Visualized Experiments. 2019 (143), 1-13 (2019).
- Leigh, W. A., et al. A high throughput machine-learning driven analysis of Ca2+ spatio-temporal maps. Cell Calcium. 91, 102260 (2020).

