Detection of SARS-CoV-2 Neutralizing Antibodies using High-Throughput Fluorescent Imaging of Pseudovirus Infection
Summary
The protocol described here outlines a fast and effective method for measuring neutralizing antibodies against the SARS-CoV-2 spike protein by evaluating the ability of convalescent serum samples to inhibit infection by an enhanced green fluorescent protein-labeled vesicular stomatitis virus pseudotyped with spike glycoprotein.
Abstract
As the COVID-19 pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to evolve, it has become evident that the presence of neutralizing antibodies against the virus may provide protection against future infection. Thus, as the creation and translation of effective COVID-19 vaccines continues at an unprecedented speed, the development of fast and effective methods to measure neutralizing antibodies against SARS-CoV-2 will become increasingly important to determine long-term protection against infection for both previously infected and immunized individuals. This paper describes a high-throughput protocol using vesicular stomatitis virus (VSV) pseudotyped with the SARS-CoV-2 spike protein to measure the presence of neutralizing antibodies in convalescent serum from patients who have recently recovered from COVID-19. The use of a replicating pseudotyped virus eliminates the necessity for a containment level 3 facility required for SARS-CoV-2 handling, making this protocol accessible to virtually any containment level 2 lab. The use of a 96-well format allows for many samples to be run at the same time with a short turnaround time of 24 h.
Introduction
In December 2019, a novel coronavirus was identified, which we now know as SARS-CoV-2, the causative agent of coronavirus disease 2019 (COVID-19)1. SARS-CoV-2 is a betacoronavirus belonging to the Coronaviridae family. These enveloped viruses comprise a large positive-sense RNA genome and are responsible for respiratory and intestinal infections in both humans and animals2. As of May 2021 there have been more than 157 million reported cases of COVID-19 globally and more than 3.2 million deaths3. The development of an effective vaccine has become the primary goal of researchers around the globe with at least 77 preclinical vaccines under investigation and 90 currently undergoing clinical trials4.
Coronaviruses encode four structural proteins including the spike protein (S), nucleocapsid (N), envelope protein (E), and the membrane protein (M). Entry of SARS-CoV-2 requires interaction of the receptor-binding domain (RBD) of S with the host receptor, human angiotensin-converting enzyme 2 (hACE2), and subsequent membrane fusion following proteolytic cleavage by host cellular serine protease, transmembrane protease serine 2 (TMPRSS2)5,6,7,8,9,10. Humoral immunodominance of the S protein of SARS-CoV has been previously reported and has now been shown also for SARS-CoV-211,12,13. Indeed, neutralizing antibody responses against S have been detected in convalescent serum from SARS-CoV patients 24 months after infection14, highlighting their critical role in the long-term immune response. The S protein has been identified as a promising vaccine target and has thus become a key component of most vaccines under development15,16.
While the rapid detection of neutralizing antibodies is a critical aspect of vaccine development, it may also shed light on the rate of infection and sero-epidemiologic surveillance in impacted areas17. A replication-competent VSV pseudotyped with the SARS-CoV-2 S glycoprotein, in place of the wild-type VSV glycoprotein, to study SARS-CoV-2 infection in biosafety level 2 settings was kindly donated by Whelan and co-workers18. VSV expressing spike (VSV-S) will be utilized to determine the neutralizing antibody response against SARS-CoV-2 spike protein. As the VSV-S used here also expresses enhanced green fluorescent protein (eGFP), eGFP foci may be detected within 24 h to quantify infection, whereas plaque formation can take 48 to 72 h. Summarized here is a simple and effective protocol to determine the ability of convalescent patient serum to neutralize VSV-S-eGFP infection. This method may also be easily adapted to interrogate other potential therapeutics that aim to disrupt the host-viral interaction of SARS-CoV-2 S protein.
Protocol
1. Plating cells (Day 1) for the production and quantification of SARS-CoV-2 pseudovirus
- Preparation for tissue culture
- Warm 1x Dulbecco's Phosphate-Buffered Saline (DPBS); Dulbecco's Modified Eagle Medium (DMEM) containing 10% Fetal Bovine Serum (FBS) and 1% penicillin/streptomycin (optional); and 0.25% trypsin-ethylenediamine tetraacetic acid (EDTA) to 37 °C in a water bath for approximately 15 min.
- Disinfect a tissue culture hood with 70% ethanol, and place tissue culture dishes, Pasteur pipettes, and serological pipettes in the tissue culture hood as needed. Remove PBS, DMEM, and trypsin from the water bath, and disinfect with 70% ethanol prior to placing in the tissue culture hood.
- Plating and maintaining cells
- Grow Vero E6 cells in DMEM (containing 10% FBS) in T75 tissue culture flasks or 15 cm tissue culture dishes in a 37 °C incubator with 5% CO2. Passage the cells as needed when cells are 80-90% confluent.
- Wash the cells by adding 10 mL of 1x PBS to each dish and gently rocking 4-5 times; aspirate the PBS. Add 3 mL of trypsin-EDTA to each dish, rock the plate to ensure the entire surface is covered. Incubate the cells at 37 °C with 5% CO2 for approximately 5 min, or until the cells have detached.
- Add 7 mL of DMEM (containing 10% FBS) to deactivate the trypsin, and resuspend the cells by pipetting up and down several times. Spin down the cells to remove the trypsin using a benchtop centrifuge at 500 × g for 5 min, aspirate the media without disturbing the cell pellet, and resuspend the cells in 10 mL of DMEM (containing 10% FBS). If cells are required for future experiments, place 1 mL in a new dish containing 15 mL of fresh DMEM (containing 10% FBS).
- Count the cells using an automated cell counter or hemocytometer, and add approximately 1 × 107 cells to each 15 cm plate, and incubate at 37 °C with 5% CO2 until they are 100% confluent (1-2 days).
2. VSV-S-EGFP pseudovirus preparation
- Infection
- Infect cells at multiplicity of infection (MOI) 0.01 with VSV-S-eGFP stock virus in 12 mL of serum-free DMEM for 1 h at 37 °C with 5% CO2, occasionally rocking the plates. Replace the inoculum with fresh DMEM (containing 2% FBS and 20 mM HEPES, pH 7.7), and move to an incubator set to 34 °C with 5% CO2.
NOTE: A temperature of 34 °C is required for the propagation of VSV-S-eGFP in cell culture. - Collect cell supernatants upon observation of extensive cytopathic effect (CPE) and cell detachment, approximately 48 h post-infection. Use a fluorescent microscope to visualize the extensive expression of eGFP by infected cells starting at 24 h post-infection.
- Infect cells at multiplicity of infection (MOI) 0.01 with VSV-S-eGFP stock virus in 12 mL of serum-free DMEM for 1 h at 37 °C with 5% CO2, occasionally rocking the plates. Replace the inoculum with fresh DMEM (containing 2% FBS and 20 mM HEPES, pH 7.7), and move to an incubator set to 34 °C with 5% CO2.
- Collection
- Centrifuge the supernatants using a benchtop centrifuge for 5 min at 1,000 × g at 4 °C to remove the cell debris. Aliquot the supernatant to avoid multiple freeze-thaw cycles, and store at -80 °C.
3. Titering the VSV-S-eGFP pseudovirus
- Serial dilution to determine viral titer
- Plate Vero E6 cells on 6-well plates at a seeding density of 6 × 105 cells per well in DMEM (with 10% FBS), and incubate overnight at 37 °C with 5% CO2.
NOTE: Vero cells that overexpress TMPRSS2 and hACE2 can also be used. - Set up a 10-fold serial dilution series of the VSV-S-eGFP virus in cold serum-free DMEM by placing 900 µL of media in seven microcentrifuge tubes. Label tubes from 1 to 7. Add 100 µL of the viral stock to the first tube and vortex briefly. Transfer 100 µL of diluted virus from tube 1 to tube 2 and vortex briefly; continue until tube 7.
- Aspirate the medium from all wells of the 6-well plate containing Vero E6 cells, and replace with 500 µL of dilutions 10-2 to 10-7. Incubate the plates for 45 min at 37 °C with 5% CO2, gently rocking every 15 min.
- Aspirate the inoculum and replace with overlay containing a 1:1 mixture of 2x DMEM and 6% carboxymethyl cellulose (CMC), final concentration of 3% CMC, supplemented with 10% FBS; incubate at 34 °C with 5% CO2 for 48 h.
NOTE: Pre-warm the overlay mixture in a 37 °C water bath for 15 min during the infection step. A 1:1 mixture of 1% agarose with 2x DMEM supplemented with 10% FBS may be used in place of CMC. To overlay with agarose, boil 1% agarose (in dH2O) and mix with cold 2x DMEM. Ensure the mixture is a suitable temperature before adding to the cell monolayer (approximately 37-40 °C). If agarose is used for the overlay, cells must be fixed by incubating in 2 mL of 3:1 methanol:acetic acid for at least one hour prior to staining.
- Plate Vero E6 cells on 6-well plates at a seeding density of 6 × 105 cells per well in DMEM (with 10% FBS), and incubate overnight at 37 °C with 5% CO2.
- Staining and calculating viral titer
- Visualize plaques by staining with crystal violet or direct visualization of eGFP under a fluorescent microscope.
- To stain plates, aspirate the overlay, wash once with PBS, then add 2 mL of 0.1% crystal violet (in 80% methanol and dH2O) to each well. Place on a plate rocker for approximately 20 min at room temperature prior to de-staining. Remove the crystal violet, and gently wash each well twice using dH2O or PBS.
NOTE: Washing and staining steps should be performed gently by pipetting slowly towards the side of each well to ensure the cell monolayer remains intact throughout the procedure. - Allow plates to dry for at least 1 h prior to counting plaques. Ensure that plaque counts are obtained from wells containing 20 to 200 plaques. To calculate the titer of the virus in plaque-forming units (PFU) per mL, multiply the dilution factor of the well counted with the volume of infection, and divide this number from the plaques present in the respective well.
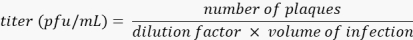
4. Plating cells (Day 1) for the measurement of neutralization of SARS-CoV-2 pseudovirus by commercially available antibodies and convalescent patient serum
- Preparation for tissue culture
- Prepare medium and tissue culture hood as described in section 1. In addition, prepare the necessary number of 96 well plates to accommodate a minimum of 2 replicates per sample (i.e., 1 plate per 6 samples); add additional replicates if sample volume permits.
- Plating and maintaining cells
- Grow and maintain Vero E6 cells as described in section 1. Prepare at least 10 mL of cell suspension per 96-well plate at a concentration of 2 × 105 cells per mL (plate 1.5 × 105 cells per mL for assays performed after 48 h, if needed). Add 100 µL of cell suspension to each well of a 96-well plate using a multichannel pipette, and incubate for 24 h at 37 °C with 5% CO2.
NOTE: Mix the cell suspension well, and rock each plate gently in all directions to ensure even distribution of cells.
- Grow and maintain Vero E6 cells as described in section 1. Prepare at least 10 mL of cell suspension per 96-well plate at a concentration of 2 × 105 cells per mL (plate 1.5 × 105 cells per mL for assays performed after 48 h, if needed). Add 100 µL of cell suspension to each well of a 96-well plate using a multichannel pipette, and incubate for 24 h at 37 °C with 5% CO2.
5. Antibody or serum dilutions and infections (Day 2)
NOTE: This protocol can be applied to measure the neutralization of VSV-S-eGFP by both commercially available antibodies and patient serum, as well as serum collected from animals for pre-clinical vaccine development studies. *Take note of the additional steps listed when handling patient/animal serum samples.
- Serial dilution series
- Prior to diluting serum samples, heat-inactivate each sample in a 56 °C water bath for 30 min to inactivate complement. Ensure necessary safety precautions are taken when handling patient samples; only open sample containers when in the tissue culture hood, and ensure appropriate containment level 2 personal protective equipment is worn.
- Set up the dilution series in an empty 96-well plate (Plate 1).
- Begin all dilutions in Row A of the 96-well plate in 80 µL. *For patient samples, begin the dilution series at 1 in 10 by placing 8 µL of serum in 72 µL of serum-free DMEM (with 1% penicillin/streptomycin).
NOTE: The concentration of neutralizing antibody used will depend on the activity predicted by the manufacturer. For the SARS-CoV-2 Spike neutralizing antibody used here (see the Table of Materials), start with 5 µg/mL to observe at least 50% neutralization. - Add 40 µL of of serum-free DMEM (with 1% penicillin/streptomycin) to rows B to G, and 80 µL to row H. Do not add virus to row H, as it serves as a cell-only control.
- Using a 12-well multichannel pipette, mix and transfer 40 µL of diluted serum or antibody from row A to row B. Mix and repeat until row F, discard 40 µL from row F. Do not add serum to row G as it represents the virus-only control row.
- Infection and overlay
- Prepare an appropriate volume of diluted VSV-S-eGFP to treat each well at MOI 0.05 (or 2000 pfu). (When completing this calculation, keep in mind that only 60 µL of the total 80 µL volume will be moved onto the cells; therefore, be sure to multiply the volume of the virus by 1.33 to maintain 2000 pfu).
NOTE: As VSV-S-eGFP is temperature-sensitive, ensure viral stocks remain on ice whenever not in use, and avoid multiple freeze-thaw cycles as titers will differ after repeated freezing. - Add 40 µL of diluted virus to each well in rows A to G of Plate 1 and mix by pipetting up and down 4-5 times. Incubate the plate for 1 h at 37 °C with 5% CO2.
- Remove the plate containing cells from the incubator (Plate 2), and carefully aspirate the media from all wells. Transfer 60 µL of antibody/virus mixture from Plate 1 to Plate 2, and incubate for 1 h at 37 °C with 5% CO2, rocking the plate every 20 min.
- Top up each well with 140 µL of overlay containing CMC in DMEM for a final concentration of 3% CMC (with 10% FBS and 1% penicillin/streptomycin). Place Plate 2 in an incubator set to 34 °C with 5% CO2 for approximately 24 h before imaging.
NOTE: Pre-warm the overlay mixture in a 37 °C water bath for 15 min during the infection step.
- Prepare an appropriate volume of diluted VSV-S-eGFP to treat each well at MOI 0.05 (or 2000 pfu). (When completing this calculation, keep in mind that only 60 µL of the total 80 µL volume will be moved onto the cells; therefore, be sure to multiply the volume of the virus by 1.33 to maintain 2000 pfu).
6. Imaging and quantification (Day 3)
- Image plates using an automated fluorescent imager (using a fluorescein isothiocyanate (FITC) filter or an alternative filter with an excitation wavelength of 488 nm). Quantify viral infection by creating a protocol that automatically identifies and counts individual eGFP foci. If an automatic counting feature is not available, use ImageJ software to quantify the number of eGFP foci.
Representative Results
This protocol outlines a rapid and effective method for detecting neutralizing antibodies against SARS-CoV-2 S protein via inhibition of VSV-S-eGFP pseudovirus infection (quantifiable by loss of eGFP foci detected). A schematic representation of the protocol is depicted in Figure 1. It is recommended that a commercially available antibody be used as a positive control each time the assay is run to ensure the consistency of the assay. Here, we demonstrate a dilution curve using a commercially available neutralizing IgG antibody against SARS-CoV-2 Spike RBD compared to an IgG control (see the Table of Materials for details about both antibodies; Figure 2).
Convalescent patient samples were collected approximately three months post SARS-CoV-2 infection, and pseudovirus neutralization was determined using the method described above (Figure 3). Importantly, minimal background inhibition was observed using healthy donor serum. Furthermore, of note, this assay distinguishes between patients with high symptom severity versus those with milder disease based on the need for hospitalization. In line with other reports, we have observed within our small cohort that hospitalized patients tend to demonstrate increased neutralizing capacity than those who did not require hospitalization19,20. While this may not always be the case for hospitalized versus non-hospitalized patient samples, the ability of the assay to differentiate varying degrees of neutralization is an asset. There may also be cases in which the neutralizing ability of the patient serum is much higher than those demonstrated here. If necessary, the starting dilution of patient serum may be adjusted, or additional dilution steps may be carried out on an additional plate.
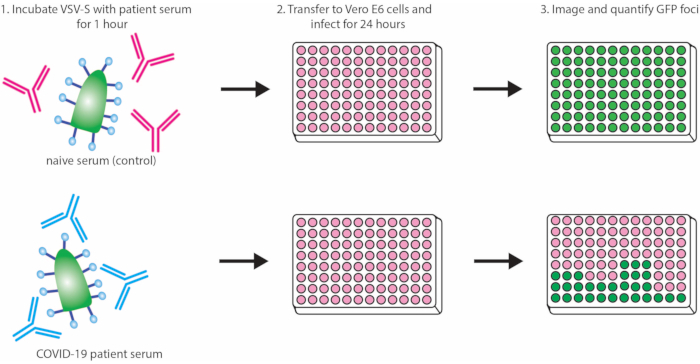
Figure 1: Protocol outline demonstrating neutralization of VSV-S-eGFP by convalescent serum. Abbreviations: VSV = vesicular stomatitis virus; eGFP = enhanced green fluorescent protein; COVID-19 = coronavirus disease. Please click here to view a larger version of this figure.
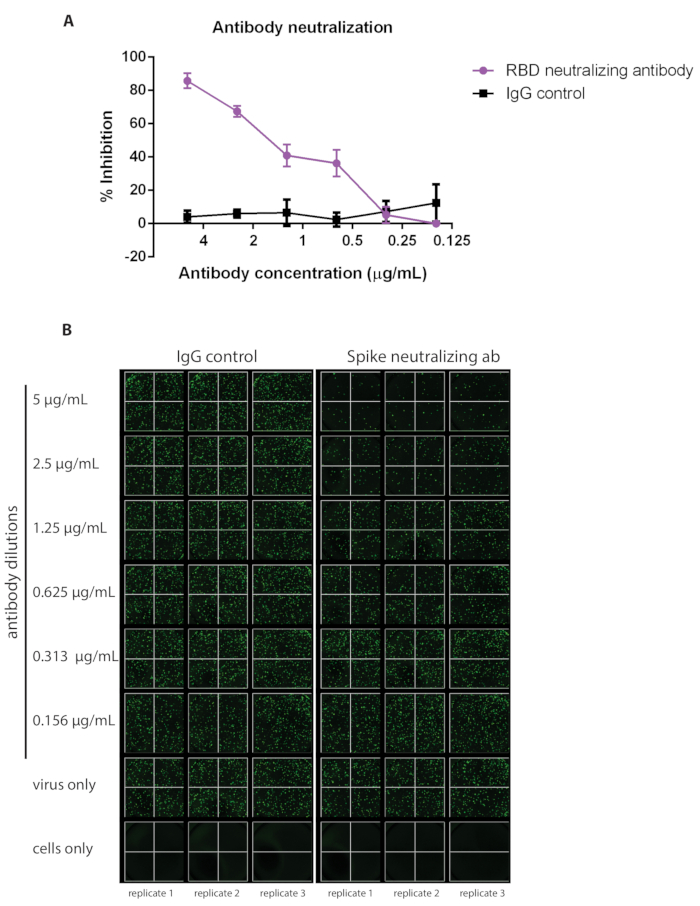
Figure 2: A commercially available neutralizing antibody against SARS-CoV-2 has been used as an example of a positive control alongside IgG as a negative control. The ability of neutralizing antibodies to inhibit viral infection will vary; refer to the information available for the specific antibody purchased to determine which concentration to begin the dilution curve (samples were run in triplicate, error bars represent ± SD). (A) Percent inhibition has been calculated based on the number of eGFP foci detected via (B) fluorescent imaging. Abbreviations: SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; IgG = immunoglobulin; SD = standard deviation; eGFP = enhanced green fluorescent protein; RBD = receptor-binding domain. Please click here to view a larger version of this figure.
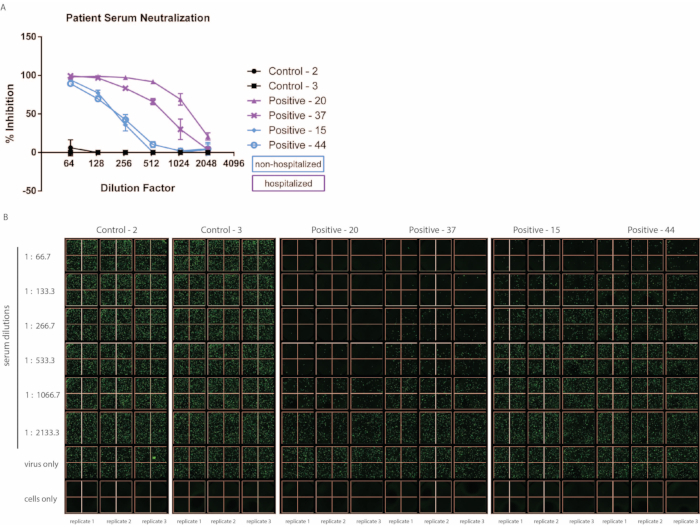
Figure 3: The ability of convalescent serum from patients to neutralize VSV-S-eGFP varies depending on the severity of symptoms. Patient serum samples were collected approximately 3 months post-SARS-CoV-2 infection, and control samples were collected from uninfected patients (samples were run in triplicate, error bars represent ± SD). (A) Percent inhibition has been calculated based on the number of eGFP foci detected via (B) fluorescent imaging. Abbreviations: SARS-CoV-2 = severe acute respiratory syndrome coronavirus 2; SD = standard deviation; eGFP = enhanced green fluorescent protein. Please click here to view a larger version of this figure.
Discussion
The method described here may be adapted to suit varying lab environments and resources as needed. Importantly, the main limitation of this protocol is the necessity for a containment level 2 space and tissue culture hood. The application of a replicating RNA virus pseudotyped with the SARS-CoV-2 spike, such as VSV-S-eGFP, is a formidable alternative to the SARS-CoV-2 virus, which requires a containment level 3 working area, but may remain a limitation for some groups. All other steps described here are quite flexible and may be performed in nearly any containment level 2 lab. For example, the use of a fluorescent imager with an automated counting feature is not necessary. The 96-well format used here means that only approximately 4 fields of view are required to image nearly the entire well using the lowest objective available (2x).
A manual fluorescent microscope may be used to image multiple fields of each well, and ImageJ (free software) can then be used to quantify eGFP foci. Additionally, we have chosen to use the 96-well format to use the lowest volume of serum possible as well as keeping consumable usage to a minimum. If a fluorescent microscope is unavailable, this protocol may be scaled up to a 6- or 12-well format to detect plaque formation after crystal violet fixation and staining (similar to the viral titer protocol described above). The most critical consideration to keep in mind when performing this protocol is the temperature sensitivity of VSV-S-eGFP. When working with VSV-S-eGFP, always aliquot the virus into small volumes to avoid multiple freeze-thaw cycles, and keep the virus on ice whenever possible. Additionally, robust fluorescent signal may be detected after 24 h; however, we have acquired similar results after imaging at 20 to 28 h post infection.
There are several methods available to detect the presence of antibodies against SARS-CoV-2, including the gold-standard enzyme-linked immunosorbent assay (ELISA) assay, which quantifies the total amount of antibodies against S21,22. Here, we have outlined a fast and reliable method to specifically detect neutralizing antibodies against the immunodominant SARS-CoV-2 spike protein in convalescent serum from patients. We have improved the classical plaque-reduction neutralization test (PRNT) by successfully adapting to a 96-well format, which allows for the detection of neutralizing antibodies in a large number of samples within 24 h by automated quantification of pseudovirus infection, allowing for quick turnaround of final data reports. In addition to detecting neutralizing antibodies within serum, this method can be adapted to perform high-throughput screening of other therapeutic strategies, which aim to directly inhibit the SARS-CoV-2 spike protein interaction with hACE2, such as monoclonal antibodies, recombinant soluble hACE2, or protease inhibitors, to impede viral entry23. With the emergence of several SARS-CoV-2 spike protein variants, it is also important to note that this method may be applied to determine if similar neutralization levels occur following infection with different variants of the virus24.
Other examples of available methods to detect antibodies against SARS-CoV-2 include ELISAs, lentivirus-based assays, and commercial kits to evaluate the neutralizing capacity of serum samples. While the commonly used IgM- or IgG-binding ELISAs are an effective method to determine the presence of antibody concentration to track previous infection or immunization, they are unable to distinguish the binding antibodies’ neutralizing capacity25. Pseudotyped lentivirus-based neutralization assays have an improved safety profile, by using non-replicative viral particles as opposed to replicating VSV-S-eGFP; however, this creates a barrier in terms of testing capacity as lentiviral titers tend to be much lower26,27. There are several commercially available kits that measure the ability of antibodies to block infection via competitive inhibition of the SARS-CoV-2 spike protein (RBD specifically) binding with the hACE2 receptor following incubation with convalescent serum (e.g., CUSABIO, GenScript, Abnova). While many of these kits have reputable sensitivity and specificity, they also tend to be relatively expensive and therefore not ideal for a large volume of samples. The protocol provided here is fast, reliable, and inexpensive. This high-throughput method may be used to test many samples, achieving a robust readout within 24 h.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to thank the Whelan lab for generously providing the VSV-S-eGFP virus used in this protocol (described in Case et al. 2020). We also thank Drs. Bill Cameron and Juthaporn Cowan (and team) for collecting the patient blood samples (REB protocol ID 20200371-01H). The authors disclose receipt of the following financial support for the research, authorship, and/or publication of this article: This work was funded by the generous support from the Ottawa Hospital Foundation and a grant from the Canadian Institutes of Health Research (#448323) and a Fast Grant from the Thistledown foundation for COVID-19 Science to C.S.I. T.R.J. is funded by an Ontario Graduate Scholarship and cluster Mitacs fellowship. JP is funded by a cluster Mitacs fellowship. T.A. is funded by a CIHR Banting Fellowship. We would also like to thank all the individuals who participated and donated their blood samples for this study.
Materials
| 0.25% trypsin-EDTA (Gibco) | Fisher scientific | LS25200114 | |
| ArrayScan VTI HCS | Thermo Fisher Scientific | Automated fluorescent imager | |
| carboxymethyl cellulose | Sigma | C5678 | |
| Dulbecco's modified Eagle's medium (Gibco) | Fisher scientific | 10-013-CV | |
| Dulbecco's modified Eagle's medium (Powder) (Gibco) | Thermo Fisher Scientific | 12-800-017 | |
| Dulbecco’s Phosphate-Buffered Saline (DPBS) | Fisher scientific | 21-031-CV | |
| HEPES | Fisher scientific | BP-310-500 | |
| IgG Isotype Control (mouse) | Thermo Fisher Scientific | 31903 | |
| Penicillin/streptomycin | Thermo Fisher Scientific | 15070063 | |
| SARS-CoV-2 (2019-nCoV) Spike Neutralizing Antibody, Mouse Mab | SinoBiological | 40592-MM57 | |
| Vero E6 cells | ATCC | CRL-1586 |
References
- Hu, B., Guo, H., Zhou, P., Shi, Z. L. Characteristics of SARS-CoV-2 and COVID-19. Nature Reviews Microbiology. 19 (3), 141-154 (2021).
- Burrell, C. J., Howard, C. R., Murphy, F. A. Coronaviruses. Fenner and White’s Medical Virlogy. , 437-446 (2017).
- COVID-19 Map. Johns Hopkins Coronavirus Resource Center Available from: https://coronavirus.jhu.edu/map.html (2021)
- Covid-19 vaccine tracker. The New York Times Available from: https://www.nytimes.com/interactive/2020/science/coronavirus-vaccine-tracker.html (2021)
- Hoffmann, M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 181 (2), 271-280 (2020).
- Letko, M., Marzi, A., Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiology. 5, 562-569 (2020).
- Azad, T., et al. Nanoluciferase complementation-based bioreporter reveals the importance of N-linked glycosylation of SARS-CoV-2 Spike for viral entry. Molecular Therapy. , (2021).
- Brown, E. E. F., et al. Characterization of critical determinants of ACE2-SARS CoV-2 RBD interaction. International Journal of Molecular Sciences. 22 (5), 2268 (2021).
- Azad, T., et al. SARS-CoV-2 S1 NanoBiT: a Nanoluciferase complementation-based biosensor to rapidly probe SARS-CoV-2 receptor recognition. Biosensors and Bioelectronics. 180, 113122 (2021).
- Azad, T., et al. Implications for SARS-CoV-2 vaccine design: Fusion of Spike glycoprotein transmembrane domain to receptor-binding domain induces trimerization. Membranes. 10 (9), 215 (2020).
- Cao, Z., et al. Potent and persistent antibody responses against the receptor-binding domain of SARS-CoV spike protein in recovered patients. Virology Journal. 7, 299 (2010).
- To, K. K. W. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. The Lancet Infectious Diseases. 20 (5), 565-574 (2020).
- Gao, Q., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 369 (6499), 77-81 (2020).
- Liu, W., et al. Two-year prospective study of the humoral immune response of patients with severe acute respiratory syndrome. Journal of Infectious Diseases. 193 (6), 792-795 (2006).
- Dong, Y., et al. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduction and Targeted Therapy. 5 (1), 237 (2020).
- Amanat, F., Krammer, F. SARS-CoV-2 vaccines: Status report. Immunity. 52 (4), 583-589 (2020).
- Amanat, F., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nature Medicine. 26 (7), 1033-1036 (2020).
- Case, J. B., et al. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host and Microbe. 28 (3), 475-485 (2020).
- Garcia-Beltran, W. F., et al. Journal Pre-proof COVID-19 neutralizing antibodies predict disease severity and survival. Cell. 184 (2), 476-488 (2020).
- Zeng, C., et al. Neutralizing antibody against SARS-CoV-2 spike in COVID-19 patients, health care workers, and convalescent plasma donors. JCI insight. 5 (22), (2020).
- Whitman, J. D., et al. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nature Biotechnology. 38 (10), 1174-1183 (2020).
- Ainsworth, M., et al. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. The Lancet Infectious Diseases. 20 (12), 1390-1400 (2020).
- Sharifkashani, S., et al. Angiotensin-converting enzyme 2 (ACE2) receptor and SARS-CoV-2: Potential therapeutic targeting. European Journal of Pharmacology. 884, 173455 (2020).
- Burki, T. Understanding variants of SARS-CoV-2. The Lancet. 397 (10273), 462 (2021).
- Jayamohan, H., et al. SARS-CoV-2 pandemic: a review of molecular diagnostic tools including sample collection and commercial response with associated advantages and limitations. Analytical and Bioanalytical Chemistry. 413 (1), 49-71 (2020).
- Nie, J., et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nature Protocols. 15 (11), 3699-3715 (2020).
- Crawford, K. H. D., et al. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 12 (5), 513 (2020).

