Tyramide Signal Amplification for the Immunofluorescent Staining of ZBP1-Dependent Phosphorylation of RIPK3 and MLKL After HSV-1 Infection in Human Cells
Summary
Tyramide signal amplification during immunofluorescent staining enables the sensitive detection of phosphorylated RIPK3 and MLKL during ZBP1-induced necroptosis after HSV-1 infection.
Abstract
The kinase Receptor-interacting serine/threonine protein kinase 3 (RIPK3) and its substrate mixed lineage kinase domain-like (MLKL) are critical regulators of necroptosis, an inflammatory form of cell death with important antiviral functions. Autophosphorylation of RIPK3 induces phosphorylation and activation of the pore-forming executioner protein of necroptosis MLKL. Trafficking and oligomerization of phosphorylated MLKL at the cell membrane results in cell lysis, characteristic of necroptotic cell death. The nucleic acid sensor ZBP1 is activated by binding to left-handed Z-form double-stranded RNA (Z-RNA) after infection with RNA and DNA viruses. ZBP1 activation restricts virus infection by inducing regulated cell death, including necroptosis, of infected host cells. Immunofluorescence microscopy permits the visualization of different signaling steps downstream of ZBP1-mediated necroptosis on a per-cell basis. However, the sensitivity of standard fluorescence microscopy, using current commercially available phospho-specific antibodies against human RIPK3 and MLKL, precludes reproducible imaging of these markers. Here, we describe an optimized staining procedure for serine (S) phosphorylated RIPK3 (S227) and MLKL (S358) in human HT-29 cells infected with herpes simplex virus 1 (HSV-1). The inclusion of a tyramide signal amplification (TSA) step in the immunofluorescent staining protocol allows the specific detection of S227 phosphorylated RIPK3. Moreover, TSA greatly increases the sensitivity of the detection of S358 phosphorylated MLKL. Together, this method enables the visualization of these two critical signaling events during the induction of ZBP1-induced necroptosis.
Introduction
Receptor-interacting serine/threonine protein kinase 3 (RIPK3) and mixed lineage kinase domain-like (MLKL) are central regulators of necroptotic cell death1,2. Necroptosis is a lytic and inflammatory form of regulated cell death involved in antiviral immunity and autoinflammation. Necroptosis of virus-infected cells immediately shuts down virus replication. Cell lysis following necroptosis induction also releases damage-associated molecular patterns, which stimulate antiviral immunity3,4. Necroptosis is initiated by the activation of RIPK3 following RIP homotypic interaction motif (RHIM)-mediated interactions with one of three upstream activating molecules: RIPK1 (upon TNF receptor 1 [TNFR1] engagement), TIR-domain-containing adapter-inducing interferon-β (TRIF; upon Toll-like receptor 3 and 4 engagement), or the antiviral nucleic acid sensor Z-DNA binding protein 1 (ZBP1)1,2. Necroptosis signaling proceeds through a series of phosphorylation events beginning with the autophosphorylation of RIPK3. The autophosphorylation of human RIPK3 at serine (S)227 inside its kinase domain is a prerequisite for necroptosis by enabling the interaction with MLKL and is commonly used as a biochemical marker for human RIPK3 activation and necroptotic cell death1,5. Once activated, RIPK3 phosphorylates the activation loop of MLKL at threonine (T)357 and S3581. This causes a change in MLKL conformation, resulting in exposure of the N-terminal four helix bundle domain. MLKL then oligomerizes and traffics to the cell membrane where it forms a pore through the insertion of the exposed four helix bundles in the lipid bilayer, eventually leading to cell death2,6.
ZBP1 is an antiviral nucleic acid sensor that recognizes left-handed Z-form nucleic acids including double-stranded RNA in the Z-conformation (Z-RNA). Z-RNA binding occurs via two Zα-domains positioned at the N-terminus of ZBP1. Z-RNA accumulating during RNA and DNA virus infection is thought to directly engage ZBP17,8. Activated ZBP1 recruits RIPK3 through its central RHIMs and induces regulated cell death, including necroptosis9,10. Viruses have adopted numerous escape mechanisms to counteract ZBP1-induced host cell necroptosis11. For example, the herpes simplex virus 1 (HSV-1) ribonucleotide reductase subunit 1, known as ICP6 and encoded by UL39, harbors RHIM at its N-terminus that interferes with ZBP1-mediated RIPK3 activation in human cells12,13,14,15. ZBP1 not only restricts viral replication, but mouse studies have shown that ZBP1 activation causes inflammatory diseases and stimulates cancer immunity16,17,18,19,20,21. Protocols that detect signaling events occurring during ZBP1-induced necroptosis in human cells are, therefore, valuable to assess the role of ZBP1 in these processes.
Tyramide signal amplification (TSA), also referred to as catalyzed reporter deposition (CARD), has been developed to improve the limit of detection and signal-to-noise ratio in antibody-based immunoassays. During TSA, any primary antibody can be used to detect the antigen of interest. Horseradish peroxidase (HRP), coupled to a secondary antibody, catalyzes the local build-up of biotinylated tyramide radicals in the presence of hydrogen peroxide. These activated biotin-tyramide radicals then react with proximal tyrosine residues to form covalent bonds. Potential tyramide-biotin substrates include the antigen itself, the primary and secondary antibodies, and neighboring proteins. Thus, while TSA significantly improves the sensitivity of the assay, some of its spatial resolution is lost. In a final step, biotin molecules are detected using fluorescently labeled streptavidin. The HRP reaction deposits many tyramide-biotin molecules on or near the antigen of interest. This greatly increases the number of streptavidin-fluorochrome binding sites, thereby greatly amplifying the sensitivity of the assay (Figure 1). Alternatively, tyramide can be directly coupled to a fluorochrome, eliminating the need for streptavidin-coupled fluorophores. Protein immunohistochemistry and DNA/RNA in situ hybridization were among the first methods whereby TSA was employed to improve signal intensities22,23. More recently, TSA has been combined with intracellular flow cytometry24 and mass spectrometry25.
Here, we present a protocol to detect serine 227 phosphorylated human RIPK3 (p-RIPK3 [S227]) and phosphorylated human MLKL (p-MLKL [S358]) upon the activation of ZBP1 by HSV-1 infection using immunofluorescence microscopy. We use a necroptosis-sensitive HT-29 human colorectal adenocarcinoma cell line that was transduced to stably express human ZBP1. These cells were infected with an HSV-1 strain expressing a mutant ICP6 protein (HSV-1 ICP6mutRHIM) in which four core amino acids within the viral RHIM (VQCG) were replaced by alanines (AAAA), thereby rendering the ICP6 unable to block ZBP1-mediated necroptosis13,14,15. To overcome the problem of the low signal-to-noise ratio of the currently commercially available antibodies directed against p-RIPK3 and p-MLKL in immunostaining26, we perform a tyramide signal amplification (TSA) step (Figure 1), which results in the robust detection of human p-RIPK3 (S227) and improves the detection sensitivity of human p-MLKL (S358) by an order of magnitude.
Protocol
1. Preparation of biotinylated tyramide
- Prepare biotinylated tyramide starting from biotin-tyramide. To make a 10 mM stock solution, dissolve 3.6 mg of biotin-tyramide in 1 mL of DMSO. Store the dissolved product in aliquots at −20 °C to preserve the quality.
2. Maintaining HT-29 cells in culture
NOTE: ZBP1-expressing HT-29 were generated by transduction with a lentivector27 encoding human ZBP1.
- Keep ZBP1-expressing HT-29 cells in McCoy's 5A medium supplemented with L-glutamine, sodium-pyruvate, and 10% fetal bovine serum (from now on referred to as full medium) and maintain in an incubator at 37 °C with 5% carbon dioxide. Ideally, use cells of a low passage number (less than 10). Leave the cells to recover for at least 5 days after the thaw cycle before the start of the experiment.
- To detach the cells from the culture flask, remove the medium and wash the cells with 5 mL of PBS (preheated to 37 °C). Next, add the appropriate amount of trypsin/EDTA (0.05% trypsin and 0.032% EDTA, respectively, preheated to 37 °C) to the cells (2 mL for a T75 flask, 3 mL for a T175 flask).
- Incubate the cells with trypsin/EDTA for up to 10 min in an incubator at 37 °C with 5% carbon dioxide. Afterward, tap the flask and visually check if the cells detached from the flask using a microscope with 4x-20x objective magnification.
- If the cells are not fully detached, incubate for another 5 min at 37 °C. When the cells are detached, stop the enzymatic reaction by adding 6 mL of full medium to the flask.
- Gather the cell suspension in a 15 mL tube. Count the cells using a trypan blue stain to assess viability; a 1:5 dilution is recommended. Only proceed with the experiment if the viability of the cells exceeds 90%.
3. Starting the experiment, seeding, and stimulation of the cells
- Seed 90,000 ZBP1-expressing HT-29 cells in full medium in a 1 cm² surface area well plate that allows high end microscopy. Use an end volume of 200 µL per well.
- Incubate the cells overnight at 37 °C with 5% carbon dioxide. Take into consideration that some extra wells need to be seeded for staining controls, which is explained in more detail in step 5.6.
- When the cells reach 70%-80% confluency, add a necroptosis-inducing stimulus to the cells. Here, the cells were infected with HSV-1 ICP6mutRHIM (multiplicity of infection [MOI] of 5, defined as plaque forming units (pfu) divided by the number of cells; the pfu was quantified using a plaque assay on Vero cells) for 6 h, 8 h, or 10 h.
- As a positive control for necroptosis induction, stimulate the cells for 4 h with a necroptosis-inducing cocktail containing 30 ng/mL TNF, 20 µM of the pan-caspase inhibitor zVAD-fmk, and 5 µM of the SMAC mimetic BV6.
- As a negative control for p-RIPK3 (S227) staining, include GSK'840 (1 µM) to inhibit RIPK3 kinase activity and prevent the autophosphorylation of S227. Ideally, add the inhibitor in an untreated condition and after necroptosis stimulation. The inhibitor can be added simultaneously with viral infection.
- Prepare the stimulations in full medium, preheated to 37 °C. Use an end volume of 200 µL per well for all stimulations.
4. Fixing the cells
- Remove the medium and wash the cells with 200 µL of 1x PBS. Then, add 150 µL of 4% PFA (already equilibrated at room temperature) to the cells and incubate at room temperature for 30 min.
NOTE: If you are using cells that detach easily, the fixation of the cells can be optimized as follows. Remove 100 µL of medium from the well, so a volume of 100 µL remains on the plate. Add 100 µL of 4% PFA to the cells. Incubate the cells for 5 min at room temperature. Afterward, remove the medium/fixative from the wells and replace with 150 µL of 4% PFA. Incubate the cells for another 20 min at room temperature to ensure complete fixation of the cells. - After fixation, remove the 4% PFA and wash the cells 3x with 200 µL of 1x PBS. The samples can be stored in an excess (>200 µL) of 1x PBS at 4 °C overnight until further processing.
5. Permeabilization and primary staining
- Remove the PBS and add 100 µL of permeabilization buffer (0.5% Triton X-100 in PBS). Incubate at room temperature for 30 min.
- Remove the permeabilization buffer and, subsequently, wash the wells with 100 µL of wash buffer (0.1% Triton X-100 in PBS). Incubate the imaging chamber with wash buffer for 5 min at room temperature on a tilting laboratory shaker (20-30 rocking motions per min).
- To prevent non-specific binding of primary antibodies, add 100 µL of blocking medium and incubate at room temperature for 2 h. Alternatively, this blocking step may be replaced by blocking with 3% BSA, 0.1% Triton-X-100 in PBS for 1 h at room temperature.
- Wash the wells 3x with 100 µL of wash buffer (0.1% Triton X-100 in PBS). Incubate the wash steps for 5 min at room temperature on a tilting laboratory shaker.
- After removing the wash buffer, add the primary antibodies to the imaging chamber and incubate overnight at 4 °C. Use an end volume of 100 µL to cover the well. During overnight incubation at 4°C, do not place the imaging chamber on a tilting laboratory shaker.
NOTE: Anti p-MLKL (S358; dilution: 1:200) and anti-p-RIPK3 (S227; dilution: 1:200) share rabbit as a host species and should, therefore, not be combined in one well. To monitor the viral infection, combine a primary antibody against a viral protein of choice with p-MLKL (S358) or p-RIPK3 (S227). Here, the cells were infected with HSV-1 infection followed by ICP0 staining (dilution: 1:50, host species: mouse). - Take into consideration the necessary staining controls at this point. Always include a no primary antibody condition to visualize the potential background of the TSA amplification step to set the masking threshold (see step 9).
NOTE: In case of a co-staining protocol (e.g., combining p-MLKL [S358] or p-RIPK3 [S227] with an antibody against a viral protein) use single stains as well. The use of single stains is important to correct for potential bleed-through signal in other imaging channels.
6. Tyramide signal amplification (TSA)
- Remove the primary antibody mix and wash wells 3x with 100 µL of wash buffer (0.1% Triton X-100 in PBS). Incubate the wash steps for 5 min at room temperature on a tilting laboratory shaker.
- Remove the wash buffer and add 100 µL of HRP-labeled secondary antibody recognizing the species of primary antibody that needs to be amplified. To amplify the p-RIPK3 (S227) or p-MLKL (S358) signal, add 100 µL of anti-rabbit-HRP and incubate for 30 min at room temperature on a tilting laboratory shaker.
NOTE: Only p-MLKL (S358) or p-RIPK3 (S227) will be amplified. The staining of a viral protein will be visualized using a standard indirect staining method. - Afterward, wash the wells 3x with 100 µL of wash buffer (0.1% Triton X-100 in PBS). Incubate the wash steps for 5 min at room temperature on a tilting laboratory shaker.
- In the next steps, add the biotinylated tyramide to the microscopic plate. Using the HRP-group coupled to the secondary antibodies, the enzymatic reaction will trigger the formation of tyramide radicals in close proximity to the primary target (here, p-MLKL [S358] or p-RIPK3 [S227]).
- To activate the enzymatic activity of HRP, add an oxidizing substrate together with the biotinylated tyramide. To achieve this, supplement the TSA buffer (0.1 M boric acid [pH 8.5]) with 0.03 M H2O2; specifically, take 5 mL of TSA buffer and add 5 µL of 30% H2O2.
- Now, dilute the biotin-tyramide in H2O2-supplemented TSA buffer ranging from 1:1,000 to 1:20,000.
NOTE: The dilution factor of the biotin-tyramide needs to be optimized per batch. - Remove the wash buffer from the wells and add diluted biotin-tyramide to the wells to an end volume of 100 µL. Incubate at room temperature for 10 min on a tilting laboratory shaker.
- Next, wash the wells 3x with 100 µL of wash buffer (0.1% Triton X-100 in PBS). Incubate the wash steps for 5 min at room temperature on a tilting laboratory shaker.
7. Fluorophores
NOTE: Since the signal of the primary antibody is converted to a biotin-group, p-MLKL (S358) and p-RIPK3 (S227) are visualized using streptavidin coupled to a fluorophore (fluorophore 568, dilution: 1:500). Additionally, the nuclei are stained with DAPI (5 µg/mL). If a viral protein is included in the staining protocol, include a suitable fluorescently labeled secondary antibody against the host species of your primary antibody. In the representative results, a mouse anti-ICP0 was used. As a secondary antibody goat anti-mouse coupled to fluorophore 633 (dilution: 1:1,000) was included.
- Make the staining mix in wash buffer (0.1% Triton X-100 in PBS) containing the antibodies and stains mentioned above. Remove the wash buffer, add 100 µL of staining mix, and incubate at room temperature for 1 h on a tilting laboratory shaker. Keep the imaging chamber shielded from light from this step onward.
- Next, wash the wells 2x with wash buffer (0.1% Triton X-100 in PBS). Incubate the wash steps for 5 min at room temperature on a tilting laboratory shaker.
- Finally, rinse the wells 2x with 1x PBS. Store the samples in an excess (> 200 µL) of 1x PBS until imaging. Alternatively, submerge the samples in mounting medium to preserve the staining. The imaging chamber is now ready to be visualized on a confocal microscope.
8. Imaging using a confocal microscope
- Turn on the confocal microscope and lasers at least 10 min before imaging.
- Use an immersion objective for sensitivity. Preferably, use 40x or 63x objective magnification. Before imaging, clean the immersion objectives with lens cleaner using appropriate cotton balls. Dirty objectives (e.g., due to dust) can result in lower quality images.
- Put an excess amount of oil on the bottom of the imaging chamber. Additionally, add a drop of oil on the objective of choice.
- Put the imaging chamber on the microscope. Find the focus field using DAPI staining or using brightfield imaging. If necessary, move around the imaging chamber to ensure proper spread of the immersion oil.
- Set up the necessary imaging tracks on the microscope. Depending on the microscope, the following steps may vary (Table 1).
NOTE: When setting up the imaging tracks, program the tracks so that the nuclear staining is measured last. The 405 nm laser may contribute to photobleaching and, thus, the loss of specific signal in all the channels. - To analyze a complete cell for the presence of p-RIPK3 (S227) or p-MLKL (S358), measure z-stacks that span the height of the cell. Here, the z-stacks were made of 40 slices every 0.16 µm, resulting in a range of 6.22 µm.
| Track | Laser | Beam splitter | Filter |
| pRIPK3 (S227) /pMLKL (S358) | 561 | MBS -405 + | BP 570-620 + LP645 |
| MBS 488/561/633 + SBS SP615 | |||
| Viral Gene: ICP0 | 633 | MBS -405 + | BP 420-480 + LP605 |
| MBS 488/561/633 + SBS LP570 | |||
| Nucleus: DAPI | 405 | MBS -405 + | BP 420-480 + BP 495-550 |
| MBS 488/561/633 |
Table 1: Imaging tracks for cell visualization.
9. Data analysis and quantification
- Upload the microscopic images to the software. Use the extended focus to visualize all the information of the z-stack in a 2D image.
- Next, program the analysis protocol in order to extract the following information: the number of cells per image, the sum of voxels showing p-RIPK3 (S227)+ or p-MLKL (S358)+ staining. A voxel represents a three-dimensional volume, defined as the three-dimensional equivalent of a pixel.
- To quantify the number of cells per image, segment the nucleus. To reduce noise in the segmentation results, add a size limitation to the segmentation (>100 µm³). The number of segmented nuclei represents the number of cells in the image.
- To quantify the amount of p-RIPK3 (S227)+ or p-MLKL (S358)+ voxels, program a threshold-based segmentation. Set the threshold so that no signal is picked up in the no primary antibody (NP) images.
- Further adapt the threshold using the images from the untreated mock condition. To ensure sensitive detection of the signal increase due to necroptosis stimulation, limit the pick-up of p-RIPK3 (S227)+ or p-MLKL (S358)+ voxels in mock conditions by setting a higher threshold. Always check the programmed analysis protocol in several images from the mock condition and necroptosis-inducing viral infection.
- Run the analysis on all image sets. Export the voxel quantification and nucleus segmentation in a .txt file for further processing in spreadsheet software.
- Make a pivot table of the data showing the image name, the nucleus quantification, and the sum of detected voxels.
- Next, divide the sum of positive voxels by the count of cells in the image. This results in a relative value of positive voxels per cell. To visualize the fold increase, divide the relative value of p-RIPK3 (S227)+ or p-MLKL (S358)+ voxels per cell by the median of the untreated condition (mock).
Representative Results
The immunofluorescent detection of MLKL phosphorylation and especially RIPK3 phosphorylation in human cells is technically challenging26. We here present an improved staining protocol for human p-RIPK3 (S227) and p-MLKL (S358) upon the activation of ZBP1. The protocol includes a TSA step to improve the detection limit and sensitivity of the fluorescent signals. To validate the method, a side-by-side comparison of the TSA-mediated immunofluorescence with standard indirect fluorescent staining of both p-RIPK3 (S227) and p-MLKL (S358) was performed.
HT-29 cells expressing human ZBP1 were infected for 9 h with an ICP6 RHIM mutant HSV-1 strain (HSV-1 ICP6mutRHIM) to induce ZBP1-mediated necroptosis and RIPK3 phosphorylation. The HSV-1 ICP6mutRHIM strain carries a VQCG to AAAA mutation within the ICP6 RHIM and is unable to block necroptosis signaling downstream of ZBP114,15. As previously reported26, standard indirect immunofluorescence was not sensitive enough to visualize RIPK3 S227 phosphorylation with the current commercially available antibody, even when the laser power of the confocal microscope was increased to 30% (Figure 2A). In contrast, the inclusion of a TSA step enabled the robust detection of p-RIPK3 (S227) in the cytosol of cells infected with HSV-1 ICP6mutRHIM. The p-RIPK3 (S227) signal reached saturation when the laser power was set at 2% (Figure 2A). Quantification of the three-dimensional z-stack images (see step 9) showed an approximate 20-fold increase in the number of voxels that were positive for p-RIPK3 (S227) in HSV-1 ICP6mutRHIM-infected over mock-treated cells (Figure 2B). Omitting the primary anti-p-RIPK3 (S227) antibody from the TSA-mediated staining protocol as a no primary (NP) control did not yield a detectable signal. To visualize HSV-1 ICP6mutRHIM-infected cells, the samples were co-stained with a primary antibody directed against the immediate early viral protein ICP0 (Figure 2A). A low but detectable p-RIPK3 (S227) signal was present in the mock-treated cells, which may represent the constitutive autophosphorylation of human RIPK3 at this site5 (see the discussion). In line with a previous report28, the RHIM of ICP6 is unable to fully block RIPK3 S227 phosphorylation, as we detected increased p-RIPK3 (S227) staining of cells infected with wild-type HSV-1 (HSV-1WT; Figure 2A,B). To further validate the specificity of the p-RIPK3 (S227) signal, the cells were treated with the RIPK3 kinase inhibitor GSK'840 prior to infection. GSK'840 binds to the kinase domain of RIPK3, preventing its activity and thereby inhibiting its autophosphorylation29. GSK'840 prevented RIPK3 phosphorylation at S227 upon ZBP1 activation (Figure 2A,B), confirming the specificity of the TSA-mediated p-RIPK3 (S227) detection method.
To follow MLKL phosphorylation, an end-stage marker for necroptosis, ZBP1-expressing HT-29 cells were infected with HSV-1 ICP6mutRHIM for 8 h and 10 h. The cells were stained with an antibody against S358 phosphorylated MLKL (p-MLKL [S358]) using TSA. The mock-treated cells showed low and somewhat punctate cytosolic staining of p-MLKL (S358), while strong p-MLKL staining was detected in the cytosol, nucleus. and at the plasma membrane in the cells that were infected with HSV-1 ICP6mutRHIM (Figure 3A). Moreover, the p-MLKL (S358) signal was observed in clusters. This is in line with the pore-forming functions of activated phosphorylated MLKL oligomers at the cell membrane and its recently reported nuclear translocation upon influenza A infection1,2,6,30. As a positive p-MLKL (S358) staining control, we stimulated ZBP1-expressing HT-29 cells with a combination of TNF, the SMAC mimetic BV6, and pan-caspase inhibitor zVAD-fmk to induce TNFR1-mediated necroptosis (Figure 3A). Omitting the primary anti-p-MLKL (S358) antibody from the TSA-mediated staining protocol as a no primary control did not yield a detectable signal.
Next, we performed a side-by-side comparison of p-MLKL (S358) immunofluorescent staining with and without TSA. ZBP1-expressing HT-29 cells were infected for 9 h with HSV-1 ICP6mutRHIM. While a laser power of 40% was needed to detect a specific p-MLKL (S358) signal in the infected cells using standard indirect immunofluorescence, the TSA-treated samples already reached saturating signals at a laser power of 6% without increasing the background staining in the mock-treated samples (Figure 3B). Moreover, quantification of the three-dimensional z-stack images showed an over 10-fold increase in the number of voxels that were positive for p-MLKL (S358) when using TSA compared to standard indirect immunofluorescence. This shows that TSA improves both the detection threshold and sensitivity for p-MLKL (S358; Figure 3C).
Finally, to validate the TSA-mediated immunofluorescence protocol for other ZBP1-dependent necroptosis viral stimuli, we infected ZBP1-expressing HT-29 cells with influenza A virus (IAV) PR8 strain for 9 h. IAV induces both ZBP1-mediated apoptosis and necroptosis in human cells30. Indeed, TSA allowed the robust detection of p-RIPK3 (S227) and p-MLKL (S358), indicative of these cells undergoing necroptosis (Figure 4A–D).
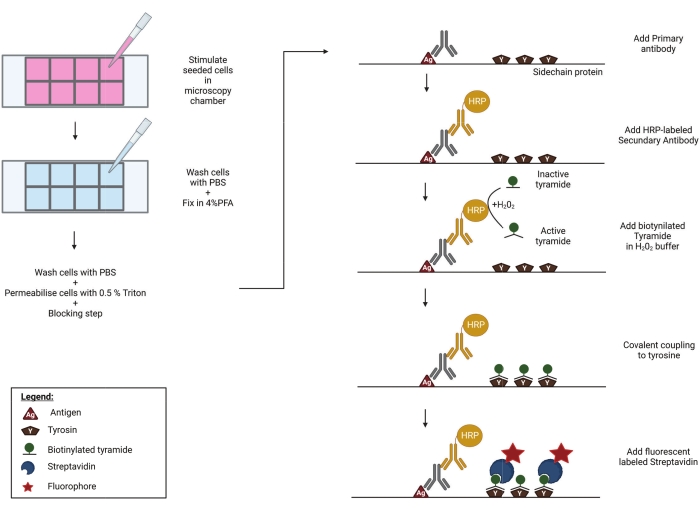
Figure 1: Schematic representation of the TSA protocol. Cells are seeded and stimulated in a well plate, compatible with high-end microscopy. Afterward, the samples are fixed in 4% PFA, permeabilised and blocked to prevent aspecific binding of primary antibodies. In order to visualize phosphorylated RIPK3 (p-RIPK3 [S227]) and MLKL (p-MLKL [S358]), specific antibodies recognizing these key phosphorylation sites are incubated overnight on the imaging chamber. Next, a secondary antibody, coupled to a horse radish peroxidase (HRP), is added. This HRP group enables the activation of biotinylated tyramide in the presence of H2O2. Subsequently, the active biotin-tyramide covalently couples to tyrosine residues in close proximity to the HRP-labeled secondary antibody. These include tyrosines on the proteins of interest-in this case p-RIPK3 or p-MLKL, as indicated in the figure-and those of neighboring proteins and on the primary and secondary antibodies themselves (not shown). This tyramide signal amplification step greatly increases the sensitivity of the staining protocol. In a final step, streptavidin coupled to a fluorescent group is added to visualize the biotinylated molecules. Please click here to view a larger version of this figure.
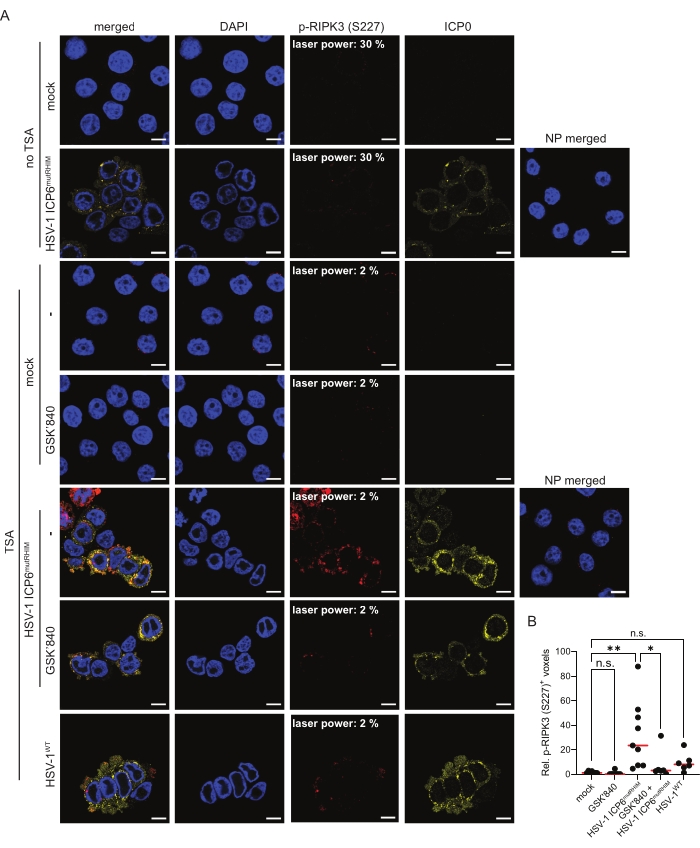
Figure 2: HSV-1 ICP6mutRHIM induces ZBP1-dependent phosphorylation of human RIPK3 at S227. (A) Representative confocal images of human ZBP1-expressing HT-29 cells, comparing TSA staining with a standard indirect (no TSA) immunofluorescence staining protocol for p-RIPK3 (S227). The mock- and virus-infected samples (HSV-1WT or HSV-1 ICP6mutRHIM [MOI = 5]) were incubated for 9 h. As a negative control, the RIPK3 kinase inhibitor GSK'840 (1 µM) was included. A no primary (NP) staining control of cells infected with HSV-1 ICP6mutRHIM (MOI = 5) for 9 h in which both the primary anti-p-RIPK3 (S227) and ICP0 antibodies were omitted is included. The laser power necessary to detect the specific p-RIPK3 (S227) signal is indicated on the images. ICP0 was used to stain the virus-infected cells, and DAPI was used to stain the nucleus. The scale bars are 10 µm. (B) Relative quantification of p-RIPK3 (S227)+ voxels using the TSA staining protocol. Every dot represents an image, and the red bar represents the median. Voxel count values are presented relative to the median of the voxel count of images in the mock condition. Statistics were done using a one-way ANOVA with multiple comparisons using Tukey correction. p > 0.05 (n.s.), p≤ 0.05 (*), p≤ 0.01 (**). Please click here to view a larger version of this figure.
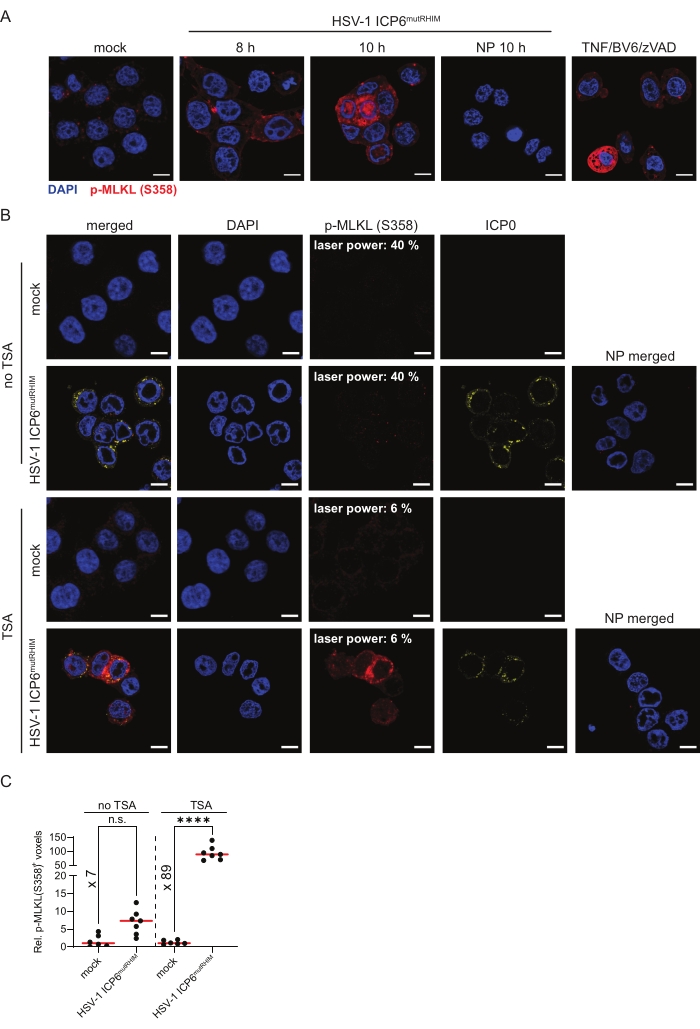
Figure 3: HSV-1 ICP6mutRHIM induces ZBP1-dependent phosphorylation of human MLKL at S358. (A) Representative confocal images of human ZBP1-expressing HT-29 cells. The cells were either mock treated or infected with HSV-1 ICP6mutRHIM (MOI = 5) for 8 h and 10 h. TSA was used to detect p-MLKL (S358). ICP0 was used to stain the virus-infected cells, and DAPI was used to stain the nucleus. A no primary (NP) staining control of cells that were infected for 10 h with HSV-1 ICP6mutRHIM (MOI = 5) in which the primary anti-p-MLKL (S358) was omitted is shown. As a positive control, the cells were stimulated for 4 h with 30 ng/mL TNF, 5 µM BV6, and 20 µM ZVAD-fmk, which induces necroptosis via TNFR1. The scale bars are 10 µm. (B) Representative confocal images of human ZBP1-expressing HT-29 cells, comparing TSA staining with a standard indirect (no TSA) immunofluorescence staining protocol for p-MLKL (S358). The cells were either mock treated or infected with HSV-1 ICP6mutRHIM (MOI = 5) for 9 h. An NP staining control of cells infected with HSV-1 ICP6mutRHIM (MOI = 5) for 9 h in which the primary anti-p-MLKL (S358) and ICP0 antibodies were omitted is included. The laser power necessary to detect the specific p-MLKL (S358) signal is indicated on the images. (C) Relative quantification of p-MLKL (S358)+ voxels using the standard (no TSA) and TSA staining protocol. Every dot represents an image, and the red bar represents the median. The voxel count values are presented relative to the median of the voxel count of images in the mock condition. Statistics were done using a one-way ANOVA with multiple comparisons using Tukey correction. p > 0.05 (n.s.), p ≤ 0.0001 (****). Please click here to view a larger version of this figure.
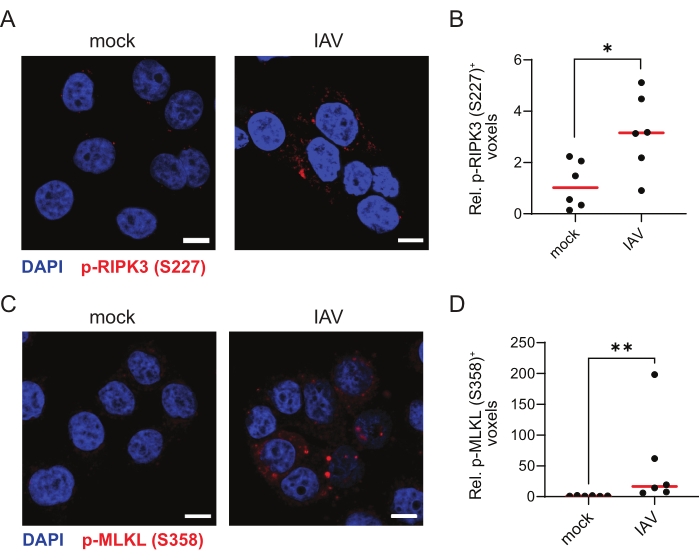
Figure 4: Influenza A virus induces ZBP1-dependent phosphorylation of human RIPK3 and MLKL. (A,C) Representative confocal images of human ZBP1-expressing HT-29 cells. The cells were either mock treated or infected with influenza A virus (IAV), PR8 strain (MOI = 4) for 9 h and stained for p-RIPK3 (S227; A) or p-MLKL (S358; C) using the TSA protocol. The scale bars are 10 µm. (B,D) Relative quantification of p-RIPK3 (S227)+ (B) or p-MLKL (S358; D) voxels. Every dot in (B,D) represents an image, and the red bar represents the median. The voxel count values are presented relative to the median of the voxel count of images in the mock condition. Statistics were done using a Mann-Whitney test. p≤ 0.05 (*), p ≤ 0.01 (**). Please click here to view a larger version of this figure.
Discussion
This immunofluorescent staining protocol describes the use of tyramide signal amplification (TSA) to increase the sensitivity for signaling events of the human necroptotic signaling pathway that are difficult to detect, including the phosphorylation of RIPK3 and MLKL26. The inclusion of a TSA step significantly improves the detection threshold of p-RIPK3 (S227) and p-MLKL (S358) and increases the sensitivity of p-MLKL (S358) straining. TSA revealed a p-RIPK3 (S227) signal already present in the mock-treated samples. In human cells, the autophosphorylation of RIPK3 at S227 is a prerequisite for necroptosis activation by enabling a stable interaction with MLKL. This process already occurs at basal levels and results in the formation of a stable inactive p-RIPK3 (S227)/MLKL dimer prior to necroptosis induction2,5,31. Similarly, the anti-p-RIPK3 antibody used in this study also detects p-RIPK3 (S227) in untreated cells by western blotting26.
The phosphorylation of MLKL by RIPK3 within the activation loop, at T357 and S358, results in dissociation of the inactive p-RIPK3 (S227)/MLKL complex and induces a conformational change whereby MLKL exposes its N-terminal four helix bundle domain. Activated p-MLKL then oligomerizes and traffics to the cell membrane where it inserts its four helix bundles into the lipid bilayer, resulting in cell lysis1,2,6. Using this TSA immunofluorescence protocol, we detected a strong increase in S358 MLKL phosphorylation during ZBP1-induced necroptosis. p-MLKL (S358) clustered within the cytosol and at the plasma membrane and was also found within the nucleus upon ZBP1 activation. Indeed, ZBP1 has been reported to stimulate MLKL-mediated perturbation of the nuclear membrane in the context of IAV infection8,30. It should be noted, however, that TSA not only deposits biotin-tyramide on the antigen of interest and the primary/secondary antibodies but also on neighboring proteins. TSA is, therefore, not ideally suited to deduce information on the precise subcellular localization of the detected proteins, and we do not recommend TSA for co-localization studies.
Repeated freeze-thaw cycles impact the stability of the biotinylated tyramide. To prevent a decrease in signal sensitivity, we recommend to aliquot the tyramide and use a fresh aliquot for each experiment. Caution should be taken with batch-to-batch differences in the biotin-tyramide stocks. If the final concentration of biotin-tyramide is too high, non-specific background of the TSA amplification will mask the specific signal. To control for this, we recommend titrating every new biotin-tyramide batch and to include a no primary staining control in which the primary antibody is omitted.
In the presented protocol, TSA was limited to one target. The detection of phosphorylated RIPK3 and MLKL was not combined in the same staining, as both primary antibodies were raised in the same species. The protocol could be adapted to detect multiple TSA-amplified signals (e.g., p-RIPK3 [S227] and p-MLKL [S358]) within the same sample using multiplex immunofluorescent TSA32,33. Finally, TSA-mediated amplification for immunofluorescence microscopy can be used for the recognition of biomarkers of other signaling pathways with reported poor signal-to-noise ratios.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to thank the VIB Bioimaging Core for training, support, and access to the instrument park. J.N. is supported by a PhD fellowship from the Research Foundation Flanders (FWO). Research in the J.M. group was supported by an Odysseus II Grant (G0H8618N), EOS INFLADIS (40007512), a junior research grant (G031022N) from the Research Foundation Flanders (FWO), a CRIG young investigator proof-of-concept grant, and by Ghent University. Research in the P.V. group was supported by EOS MODEL-IDI (30826052), EOS INFLADIS (40007512), FWO senior research grants (G.0C76.18N, G.0B71.18N, G.0B96.20N, G.0A9322N), Methusalem (BOF16/MET_V/007), iBOF20/IBF/039 ATLANTIS, Foundation against Cancer (F/2016/865, F/2020/1505), CRIG and GIGG consortia, and VIB.
Materials
| Antibodies | |||
| Anti-rabbit HRP | Agilent Technologies Belgium | K4002 | Envision+ System-HRP Labelled Polymer anti-rabbit |
| Goat anti-mouse DyLight 633 | Thermofisher | 35513 | Secundary antibody |
| HSV-1 ICP0 | Santa Cruz | sc-53070 | Mouse anti-ICP0(HSV-1) antibody |
| IAV-PR8 mouse serum | In house production | xx | Mouse anti-IAV-PR8 polyclonal antibody |
| pMLKL | Abcam | ab187091 | Rabbit anti-MLKL-phospho S358 antibody |
| pRIPK3 | Abcam | ab209384 | Rabbit anti-RIPK3-phospho S227 antibody |
| Fluorophores | |||
| DAPI | Thermofisher | D21490 | To visualise the nucleus of the cells |
| Streptavidin coupled to Alexa Fluor 568 | Thermofisher | S11226 | To visulalise biotin molecules |
| Compounds | |||
| 30% H2O2 | Sigma | H1009 | Oxidising substrate, necessary for HRP activity |
| 4% PFA | SANBIO | AR1068 | To fix/crosslink the cells |
| Biotinyl-tyramide | R&D Systems | 6241 | To amplify signal, HRP substrate |
| BV-6 | Selleckchem | S7597 | BV6 IAP Inhibitor |
| For cell culture: to detach the cells | |||
| 8.0 g/L NaCl | |||
| 0.4 g/L disodium salt of EDTA | |||
| EDTA 0.04% | In house formulation | 1.1 g/L Na2HPO4 | |
| 0.2 g/L NaH2PO4 | |||
| 0.2 g/L KCl | |||
| 0.2 g/L Glucose | |||
| Fetal Bovine serum | TICO | FBS EU XXX | For cell culture, maintaining cell culture; lot number: 90439 |
| GSK'840 | Aobious | AOB0917 | RIPK3 kinase inhibitor |
| L-Glutamine | Sigma-Aldrich | G7513 | For cell culture, maintaining cell culture |
| MAXblock | Active Motif | 15252 | Blocking solution |
| PBS | Gibco | 10444402 | |
| Sodium pyruvate | Sigma-Aldrich | S8636 | For cell culture, maintaining cell culture |
| TNF-α | In house production | – | Signaling molecule, able to trigger cell death in combination with BV6 and zVAD |
| Triton X-100 | Sigma Aldrich | T8787-50ML | To permeabilise the cells |
| Trypan blue | Merck | 11732 | For cell counting, used as live/dead marker at 0,1% |
| Trypsine | Sigma-Aldrich | T4424 | For cell culture: to detach the cells |
| zVAD | Bachem | BACE4026865.0005 | Z-Val-Ala-DL-Asp-fluoromethylketone |
| Material | |||
| µ-Slide 8 well high glass bottom | iBidi | 80807 | To culture the cells |
| Cotton Preping Balls-size medium | Electron Microscopy Sciences | 71001-10 | To clean the objectives |
| Immersol 518 F / 30 °C | ZEISS | 444970-9000-000 | To visualise the sample at high magnifications |
| Lens Cleaner | ZEISS | 000000-0105-200 | To clean the objectives |
| LSM880 Fast Airyscan confocal microscope | To visualise the sample | ||
| Software | |||
| Excel | Office | xx | To process the data |
| Prism 9 | Graphpad | xx | To analyse the data- statistical testing and graph generation |
| Volocity 6.3 | Volocity | xx | To perform quantifications |
| Zen black | ZEISS | xx | To aquire and process images |
| Zen blue | ZEISS | xx | To visualise images |
| Viruses | |||
| HSV-1 (mutRHIM) F strain | produced by Dr. Jiahuai Han | in house replication | HSV-1 as a trigger for necroptosis; RHIM core domain of UL39/ICP6 is mutated (VQCG>AAAA) |
| HSV-1 (WT) F strain | Produced by Dr. Jiahuai Han | in house replication | HSV-1 (WT) as a negative control for necroptosis induction (ICP6 inhibition) |
| IAV PR8 | in house stock | in house replication | IAV as a trigger for necroptosis |
References
- Meng, Y., Sandow, J. J., Czabotar, P. E., Murphy, J. M. The regulation of necroptosis by post-translational modifications. Cell Death & Differentiation. 28 (3), 861-883 (2021).
- Petrie, E. J., Czabotar, P. E., Murphy, J. M. The structural basis of necroptotic cell death signaling. Trends in Biochemical Sciences. 44 (1), 53-63 (2019).
- Mocarski, E. S., Guo, H., Kaiser, W. J. Necroptosis: The Trojan horse in cell autonomous antiviral host defense. Virology. 479-480, 160-166 (2015).
- Nailwal, H., Chan, F. K. Necroptosis in anti-viral inflammation. Cell Death & Differentiation. 26 (1), 4-13 (2019).
- Meng, Y., et al. Human RIPK3 maintains MLKL in an inactive conformation prior to cell death by necroptosis. Nature Communications. 12 (1), 6783 (2021).
- Samson, A. L., Garnish, S. E., Hildebrand, J. M., Murphy, J. M. Location, location, location: A compartmentalized view of TNF-induced necroptotic signaling. Science Signalling. 14 (668), (2021).
- Koehler, H., et al. Vaccinia virus E3 prevents sensing of Z-RNA to block ZBP1-dependent necroptosis. Cell Host Microbe. 29 (8), 1266-1276 (2021).
- Balachandran, S., Mocarski, E. S. Viral Z-RNA triggers ZBP1-dependent cell death. Current Opinion in Virology. 51, 134-140 (2021).
- Kuriakose, T., Kanneganti, T. D. ZBP1: Innate sensor regulating cell death and inflammation. Trends in Immunology. 39 (2), 123-134 (2018).
- Maelfait, J., Liverpool, L., Rehwinkel, J. Nucleic acid sensors and programmed cell death. Journal of Molecular Biology. 432 (2), 552-568 (2020).
- Verdonck, S., Nemegeer, J., Vandenabeele, P., Maelfait, J. Viral manipulation of host cell necroptosis and pyroptosis. Trends in Microbiology. 30 (6), 593-605 (2022).
- Guo, H., et al. Herpes simplex virus suppresses necroptosis in human cells. Cell Host & Microbe. 17 (2), 243-251 (2015).
- Yu, X., et al. Herpes simplex virus 1 (HSV-1) and HSV-2 mediate species-specific modulations of programmed necrosis through the viral ribonucleotide reductase large subunit R1. Journal of Virology. 90 (2), 1088-1095 (2016).
- Huang, Z., et al. RIP1/RIP3 binding to HSV-1 ICP6 initiates necroptosis to restrict virus propagation in mice. Cell Host & Microbe. 17 (2), 229-242 (2015).
- Guo, H., et al. Species-independent contribution of ZBP1/DAI/DLM-1-triggered necroptosis in host defense against HSV1. Cell Death & Disease. 9 (8), 816 (2018).
- Devos, M., et al. Sensing of endogenous nucleic acids by ZBP1 induces keratinocyte necroptosis and skin inflammation. The Journal of Experimental Medicine. 217 (7), 20191913 (2020).
- Jiao, H., et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature. 580 (7803), 391-395 (2020).
- de Reuver, R., et al. ADAR1 prevents autoinflammation by suppressing spontaneous ZBP1 activation. Nature. 607 (7920), 784-789 (2022).
- Jiao, H., et al. ADAR1 averts fatal type I interferon induction by ZBP1. Nature. 607 (7920), 776-783 (2022).
- Hubbard, N. W., et al. ADAR1 mutation causes ZBP1-dependent immunopathology. Nature. 607 (7920), 769-775 (2022).
- Zhang, T., et al. ADAR1 masks the cancer immunotherapeutic promise of ZBP1-driven necroptosis. Nature. 606 (7914), 594-602 (2022).
- Adams, J. C. Biotin amplification of biotin and horseradish peroxidase signals in histochemical stains. The Journal of Histochemistry and Cytochemistry. 40 (10), 1457-1463 (1992).
- Speel, E. J., Hopman, A. H., Komminoth, P. Tyramide signal amplification for DNA and mRNA in situ hybridization. Methods in Molecular Biology. 326, 33-60 (2006).
- Clutter, M. R., Heffner, G. C., Krutzik, P. O., Sachen, K. L., Nolan, G. P. Tyramide signal amplification for analysis of kinase activity by intracellular flow cytometry. Cytometry A. 77 (11), 1020-1031 (2010).
- Dopie, J., Sweredoski, M. J., Moradian, A., Belmont, A. S. Tyramide signal amplification mass spectrometry (TSA-MS) ratio identifies nuclear speckle proteins. The Journal of Cell Biology. 219 (9), 201910207 (2020).
- Samson, A. L., et al. A toolbox for imaging RIPK1, RIPK3, and MLKL in mouse and human cells. Cell Death and Differentiation. 28 (7), 2126-2144 (2021).
- De Groote, P., et al. Generation of a new gateway-compatible inducible lentiviral vector platform allowing easy derivation of co-transduced cells. Biotechniques. 60 (5), 252-259 (2016).
- Ali, M., Roback, L., Mocarski, E. S. Herpes simplex virus 1 ICP6 impedes TNF receptor 1-induced necrosome assembly during compartmentalization to detergent-resistant membrane vesicles. The Journal of Biological Chemistry. 294 (3), 991-1004 (2019).
- Mandal, P., et al. RIP3 induces apoptosis independent of pronecrotic kinase activity. Moleular Cell. 56 (4), 481-495 (2014).
- Zhang, T., et al. Influenza virus Z-RNAs induce ZBP1-mediated necroptosis. Cell. 180 (6), 1115-1129 (2020).
- Garnish, S. E., et al. Conformational interconversion of MLKL and disengagement from RIPK3 precede cell death by necroptosis. Nature Communications. 12 (1), 2211 (2021).
- Clay, H., Ramakrishnan, L. Multiplex fluorescent in situ hybridization in zebrafish embryos using tyramide signal amplification. Zebrafish. 2 (2), 105-111 (2005).
- Parra, E. R., et al. Procedural requirements and recommendations for multiplex immunofluorescence tyramide signal amplification assays to support translational oncology studies. Cancers. 12 (2), 255 (2020).

