A “Dual-Addition” Calcium Fluorescence Assay for the High-Throughput Screening of Recombinant G Protein-Coupled Receptors
Summary
In this work, a high-throughput, intracellular calcium fluorescence assay for 384-well plates to screen small molecule libraries on recombinant G protein-coupled receptors (GPCRs) is described. The target, the kinin receptor from the cattle fever tick, Rhipicephalus microplus, is expressed in CHO-K1 cells. This assay identifies agonists and antagonists using the same cells in one “dual-addition” assay.
Abstract
G protein-coupled receptors (GPCRs) represent the largest superfamily of receptors and are the targets of numerous human drugs. High-throughput screening (HTS) of random small molecule libraries against GPCRs is used by the pharmaceutical industry for target-specific drug discovery. In this study, an HTS was employed to identify novel small-molecule ligands of invertebrate-specific neuropeptide GPCRs as probes for physiological studies of vectors of deadly human and veterinary pathogens.
The invertebrate-specific kinin receptor was chosen as a target because it regulates many important physiological processes in invertebrates, including diuresis, feeding, and digestion. Furthermore, the pharmacology of many invertebrate GPCRs is poorly characterized or not characterized at all; therefore, the differential pharmacology of these groups of receptors with respect to the related GPCRs in other metazoans, especially humans, adds knowledge to the structure-activity relationships of GPCRs as a superfamily. An HTS assay was developed for cells in 384-well plates for the discovery of ligands of the kinin receptor from the cattle fever tick, or southern cattle tick, Rhipicephalus microplus. The tick kinin receptor was stably expressed in CHO-K1 cells.
The kinin receptor, when activated by endogenous kinin neuropeptides or other small molecule agonists, triggers Ca2+ release from calcium stores into the cytoplasm. This calcium fluorescence assay combined with a "dual-addition" approach can detect functional agonist and antagonist "hit" molecules in the same assay plate. Each assay was conducted using drug plates carrying an array of 320 random small molecules. A reliable Z' factor of 0.7 was obtained, and three agonist and two antagonist hit molecules were identified when the HTS was at a 2 µM final concentration. The calcium fluorescence assay reported here can be adapted to screen other GPCRs that activate the Ca2+ signaling cascade.
Introduction
G protein-coupled receptors (GPCRs), which are present from yeast to humans, represent the largest superfamily of receptors in many organisms1. They play critical roles in regulating nearly all biological processes in animals. There are 50-200 GPCRs in the genome of arthropods, meaning they represent the largest membrane receptor superfamily2. They are classified into six major classes, A-F, based on their sequence similarity and functions3. GPCRs transduce various extracellular signals, such as those of hormones, neuropeptides, biogenic amines, glutamate, proton, lipoglycoproteins, and photons4. GPCRs couple to heterotrimer G proteins (Gα, Gβ, and Gγ) to transmit downstream signals. GPCRs coupled to Gαs or Gαi/o proteins increase or decrease, respectively, the intracellular 3', 5'-cyclic adenosine monophosphate (cAMP) levels by activating or inhibiting adenylyl cyclase. GPCRs coupled to Gαq/11 induce calcium release from the endoplasmic reticulum calcium stores by activating the phospholipase C (PLC)-inositol-1,4,5-triphosphate (IP3) pathway. GPCRs coupled to Gα12/13 activate RhoGTPase nucleotide exchange factors5,6. GPCRs are the target of more than 50% of human drugs and an acaricide, amitraz4. As GPCRs transduce such diverse signals, they are promising targets for developing novel pesticides that disrupt invertebrate-specific physiological functions.
The goal of HTS is to identify hit molecules that can modulate receptor functions. HTS involves assay development, miniaturization, and automation7. Arthropod neuropeptide GPCRs are involved in most physiological functions, such as development, molting and ecdysis, excretion, energy mobilization, and reproduction4. Most of the neuropeptide GPCRs of arthropods and metazoans signal through the calcium signaling cascade2,6,8,9,10, such as in the myoinhibitory peptide and SIFamide receptors of the blacklegged tick Ixodes scapularis; their ligands are antagonistic in hindgut motility assays, with SIF eliciting contraction and MIP inhibiting it11,12. An NPY-like receptor of the yellow fever mosquito, Aedes aegypti, regulates female host seeking13. Compared to other alternative calcium mobilization assays such as the aequorin calcium bioluminescence assay14, the calcium fluorescence assay is easy to perform, does not require the transfection of other recombinant calcium detecting proteins, and is cost-effective. The calcium fluorescence assay produces a prolonged signal compared to the fast kinetic signal obtained in the aequorin calcium bioluminescence assay14,15.
In the example here, the kinin receptor from the cattle fever tick, Rhipicephalus microplus, was recombinantly expressed in the CHO-K1 cell line and used for the calcium fluorescence assay. There is only one kinin receptor gene found in R. microplus; the receptor signals through a Gq protein-dependent signaling pathway and triggers the efflux of Ca2+ from calcium stores into the intracellular space16. This process can be detected and quantified by a fluorophore, which elicits a fluorescence signal when binding calcium ions (Figure 1).
The kinin receptor is an invertebrate-specific GPCR, which belongs to the Class A Rhodopsin-like receptors. Kinin is an ancient signaling neuropeptide that is present in Mollusca, Crustacea, Insecta, and Acari4,17,18. Coleopterans (beetles) lack the kinin signaling system; in the mosquito Aedes aegypti, there is only one kinin receptor that binds three aedeskinins, while Drosophila melanogaster has one kinin receptor with drosokinin as a unique ligand19,20,21. There are no homologous kinins or kinin receptors in vertebrates. Although the exact function of kinin is unknown in ticks, the kinin receptor RNAi-silenced females of R. microplus show significantly reduced reproductive fitness22. Kinins are pleotropic peptides in insects. In Drosophila melanogaster, they are involved in both the central and peripheral nervous regulatory systems23, pre-ecdysis24, feeding25, metabolism26, and sleep activity patterns26,27, as well as larval locomotion28. Kinins regulate hindgut contraction, diuresis, and feeding in the mosquito A. aegypti29,30,31. The kinin peptides have a conserved C-terminal pentapeptide Phe-X1-X2-Trp-Gly-NH2, which is the minimum required sequence for biological activity32. The arthropod specificity, the small size of the endogenous ligand, which makes them amenable to small-molecule interference, and the pleiotropic functions in insects make the kinin receptor a promising target for pest control4.
The "dual-addition" assay (Figure 2) allows the identification of agonists or antagonists in the same HTS assay15. It is adapted from a "dual-addition" assay that is commonly used in the pharmaceutical industry for drug discovery33. In brief, the first addition of drugs into the cell plate allows the identification of potential agonists in the chemical library when a higher fluorescence signal is detected compared to the application of the solvent control. After 5 min of incubation with these small molecules, a known agonist (kinin peptide) is applied to all the wells. Those wells that randomly received an antagonist from the drug plate display a lower fluorescence signal upon agonist addition compared to the control wells that received the solvent in the first addition. This assay then allows the identification of potential agonists and antagonists with the same cells. In a standard HTS project, these hit molecules would be further validated through dose-response assays and by additional biological activity assays, which are not shown here.
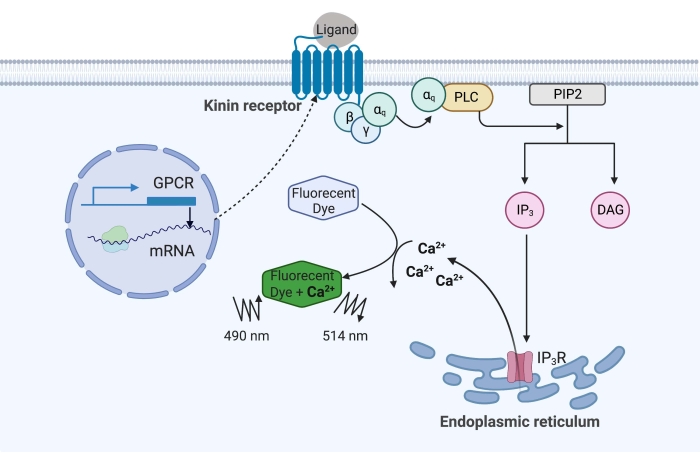
Figure 1: Illustration of the calcium fluorescence assay mechanism. The Gq protein triggers the intracellular calcium signaling pathway. The kinin receptor (G protein-coupled receptor) was recombinantly expressed in CHO-K1 cells. When the agonist ligand binds to the receptor, the Gq protein associated with the kinin receptor activates PLC, which catalyzes the conversion of a PIP2 molecule into IP3 and DAG. IP3 then binds to the IP3R on the surface of the endoplasmic reticulum, leading to the release of Ca2+ into the cytoplasm, where Ca2+ ions bind to the fluorophores and elicit a fluorescence signal. The fluorescence signal can be obtained by excitation at 490 nm and detected at 514 nm. Abbreviations: GPCR = G protein-coupled receptor; PLC = phospholipase C; PIP2 = phosphatidylinositol 4,5-bisphosphate; IP3 = inositol trisphosphate; DAG = diacylglycerol; IP3R = IP3 receptor. Created with BioRender.com. Please click here to view a larger version of this figure.
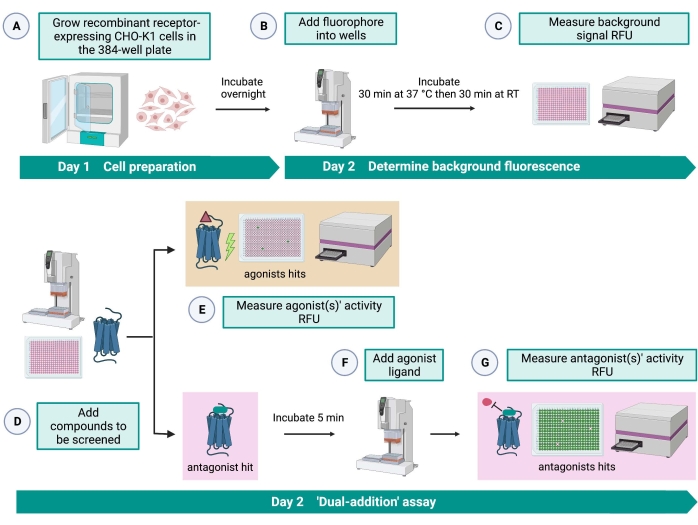
Figure 2: The workflow for the high-throughput screening of small molecules on a G protein-coupled receptor expressed in CHO-K1 cells. (A) Recombinant CHO-K1 cells stably expressing the kinin receptor were added to the 384-well plate (10,000 cells/well) using a liquid handling system (25 µL/well) and incubated in a humidified CO2 incubator for 12-16 h. (B) The assay buffer containing the fluorescent dye (25 µL/well) was added into the cell plate using a liquid handling system. The plate was incubated for 30 min at 37 °C for 30 min and equilibrated at RT for another 30 min. (C) The background fluorescence signal of the cells in each well was measured with a plate reader. (D) Drug solutions from a 384-well library plate and blank solvent (all at 0.5 µL/well) were added into the cellular assay plate using a liquid handling system. (E) Cellular calcium fluorescence responses were measured with the plate reader immediately after the addition of the drug solutions; compound(s) eliciting higher than average fluorescence signals were picked out as agonist hit(s). Antagonist hits that block the GPCR (icon below) were revealed after the addition of the peptide agonist during step G. (F) In the same assay plate, after 5 min of incubation of the cells with screening compounds, an endogenous agonist peptide Rhimi-K-1 (QFSPWGamide) of the tick kinin receptor was added to each well (1 µM). (G) Cellular fluorescence responses after the agonist peptide addition were measured by the plate reader immediately. Compound(s) inhibiting the fluorescence signal were selected as antagonist hit(s). Abbreviations: GPCR = G protein-coupled receptor; RT = room temperature; RFU = relative fluorescence units. Created with BioRender.com. Please click here to view a larger version of this figure.
Protocol
1. Cell maintenance
NOTE: A CHO-K1 cell line that stably expresses the kinin receptor from R. microplus, named BMLK3, was developed by Holmes et al.16. The details of the development of the cell line are presented elsewhere14. All the following steps are performed under sterile conditions in a class II biosafety cabinet.
- Grow the recombinant cell line in selective media (F-12K medium containing 10% fetal bovine serum [FBS] and 800 µg/mL G418 sulfate) to ensure the expression of the target receptor. Store the cells stably expressing the receptor in 1 mL of freezing medium (90% FBS and 10% dimethyl sulfoxide [DMSO]) in a 2 mL cryovial in a −80 °C freezer.
NOTE: For long-term storage of the frozen cells, store them in liquid nitrogen. - Prewarm all the media before cell culture. In a biosafety cabinet, transfer 13 mL of prewarmed selective medium (F-12K medium containing 10% FBS and 800 µg/mL G418 sulfate) into a T-75 flask, and keep it in the biosafety cabinet. Thaw one vial of the frozen BMLK3 cells (~1.5 × 106 cells) in a 37 °C water bath for 2-3 min. Transfer the thawed cells into the T-75 flask. Maintain the cells in a humidified incubator at 37 °C and 5% CO2 unless specified otherwise.
- When the cells reach 90% confluency (1-2 days), prewarm all the media to 37 °C except for Dulbecco's phosphate-buffered saline (DPBS), which is kept at room temperature. In a biosafety cabinet, remove the spent medium from the T-75 flask, wash the cells for 5 s with 10 mL of DPBS by swirling the flask gently, and then remove the DPBS with a serological pipette.
NOTE: An image of CHO-K1 cells at 90% confluency is shown in Lu et al.14. - Detach the cells from the T-75 flask by adding 2 mL of 0.25% trypsin-EDTA, and incubate for 3-5 min at 37 °C in the incubator. Then, add 8 mL of selective medium, and mix well by pipetting and releasing gently 2x-3x with the same serological pipette.
- Transfer 2 mL of the cell suspension (~1 × 106 cells) from step 1.4 into a new T-75 flask containing 10 mL of warm selective medium. Grow the cells for 1-2 days in the incubator until they reach 90% confluency.
- Use the cells for the assay following the next steps, or repeat steps 1.4-1.5 once or twice before using the cells in the assay.
NOTE: Do not exceed three to four passages, as the assay signal with certain cell lines may become weaker with further passages.
2. Calcium fluorescence assay
- Coat the cell plate.
- Use a liquid handling system placed inside a biosafety cabinet for all the pipetting steps in the 384-well plates. Create custom programs for pipetting into the 384-well plates31 (Supplemental Table S1). Coat the sterile 384-well plates in advance. In a biosafety cabinet, load 10 µL/well of an aqueous solution of Poly-D-lysine (PDL) at 0.05 mg/mL into each plate, and incubate for 5 min at room temperature.
- Empty the plate by quickly inverting the plate and gently blotting it on sterile paper towels. Then, rinse each well with 10 µL of water, empty the plate, and leave the plate to dry in the biosafety cabinet overnight without the lid. Close the plate with the lid, and store at 4 °C in the refrigerator.
NOTE: The coated plates can be stored at 4 °C for up to 6 months.
- Day 1
- Take out the drug plate (100 µM in 90% DPBS + 10% DMSO, final concentration in the well for HTS will be 2 µM) stored in the −20 °C freezer, and place it at room temperature.
NOTE: Layout of the drug plate: Each 384-well plate (24 columns x 16 rows) contains 320 wells with different library compounds and 64 wells with blank solvent (DPBS containing 10% DMSO), which are arranged in four columns, with two columns on the edge of each side. See Supplemental Table S2 for the plate layout. - When cells reach ~70%-90% confluency in the T-75 flask, detach the cells from the T-75 flask as described below. Prewarm all the media to 37 °C except for the DPBS (room temperature).
- Remove the spent medium, wash the cells with 10 mL of DPBS, and then remove the DPBS. Detach the cells from the T-75 flask using 2 mL of 0.25% trypsin-EDTA for 3-5 min at 37 °C in the incubator, add 8 mL of selective medium, and transfer the cell suspension into a 15 mL conical tube to centrifuge at 1,000 × g for 3 min.
- Discard the supernatant, and resuspend the cell pellet in 10 mL of F-12K medium containing 1% FBS and 400 µg/mL G418 sulfate. Keep the suspension in the biosafety cabinet while determining the cell count.
- Determine the cell density of the suspension for further dilution: Mix 20 µL of cell suspension into 20 µL of 0.4% trypan blue, and then load 20 µL of the mixture into a cell counting chamber to be read by a cell counter for cell density.
- Dilute the cell suspension in the same medium (F-12K medium containing 1% FBS and 400 µg/mL G418 sulfate) to a final volume of at least 15 mL at a density of 4 × 105 cells/mL.
- Seed the cells in the PDL-coated 384-well plate. Transfer 15 mL of the above cell suspension (4 × 105 cells/mL) into a 150 mL auto-friendly reagent reservoir. Dispense 25 µL of the cell suspension (~10,000 cells/well) into each well of the 384 wells of the plate using a liquid handling system in two steps. Load 384/12.5 µL low-retention tips onto the liquid handler head, aspirate 12.5 µL from the reservoir (speed 5.2 µL/s), and dispense into each well (speed: 3.1 µL/s).
- Repeat the pipetting as in previous step 2.2.5 to reach 25 µL per well. Then, incubate the plate overnight (14-16 h) at 37 °C and 5% CO2 in the humidified incubator.
NOTE: As the maximum pipetting volume is 12.5 µL for this specific head, pipetting for 25 µL is performed in two steps.
- Take out the drug plate (100 µM in 90% DPBS + 10% DMSO, final concentration in the well for HTS will be 2 µM) stored in the −20 °C freezer, and place it at room temperature.
- Day 2
- The next morning, check the covered cell plate under a microscope; if not confluent, wait until the cells reach 90% confluency.
- Prepare a stock solution of fluorescent dye: Resuspend lyophilized fluorescent dye in 100 µL of DMSO, and avoid direct light on the stock solution. Wrap the tube with aluminum foil to prevent photobleaching.
NOTE: The stock can be aliquoted into 15 µL aliquots for each plate assay to avoid repeated freezing and thawing; the aliquots can be stored at −80 °C for up to 1 month. - In the biosafety cabinet with its lights off, prepare loading dye (1x) in a 15 mL conical tube wrapped with aluminum foil, combining 15 µL of the fluorescent dye stock solution (from step 2.3.2) and the rest of the prewarmed kit components (37 °C): 13.5 mL of 1x HHBS (Hank's buffer with 20 mM HEPES) and 1.5 mL of reagent B (dye efflux inhibitor).
NOTE: The room light is on during this step. - Close the tube and mix well by gently inverting the tube several (typically 3-5x) times.
NOTE: Keep at room temperature, and use the loading dye mixture within 30 min. - When the cells reach 90% confluency, remove the spent medium from the 384-well assay plate by quickly inverting the plate and gently blotting on sterile paper towels; repeat this movement 2x-3x to remove all the liquid from the plate. Discard the wet towels.
- Turn off most artificial direct lights in the room (leaving a soft, dim desk lamp on or similar to allow visible work conditions) and in the biosafety cabinet for all the steps until the end of the assay (approximately for 1.5 h).
- Place 15 mL of the loading dye (1x) from step 2.3.3 into a 150 mL auto-friendly reservoir.
- Transfer 25 µL of the loading dye (1x) from the reservoir into each well using a liquid handling system with 384/12.5 µL low-retention tips by dispensing 12.5 µL into each well of the plate (aspiration and dispensing speed: 3.8 µL/s).
- Repeat the pipetting step as in 2.3.7.1 to reach a final volume of 25 µL in each well of the plate. Cover and wrap the plate with aluminum foil to protect it from ambient light.
- Incubate the covered cell plate at 37 °C in the CO2 humidified incubator for 30 min, remove it from the incubator, and equilibrate it at room temperature inside the plate reader or on the bench, keeping the plate covered and wrapped with foil for another 30 min. The cells are then ready for high-throughput screening (HTS).
- Chemical preparation: Spin the drug plate from step 2.2.1 at 1,200 × g in a plate centrifuge for 1 min at room temperature.
- Prepare a 10x agonist peptide solution of Rhimi-K-1 (QFSPWGamide) (10 µM) by resuspending 100 nmoles of lyophilized peptide in 10 mL of 1x HHBS containing 0.1% DMSO, and transfer the solution into a 150 mL auto-friendly reservoir.
- Measure the background signal: For the entire HTS assay in the plate reader, under Protocols, choose end-point fluorescence mode.
NOTE: The fluorescent signal is read from the bottom of the plate at excitation/emission wavelengths of 495 nm/525 nm.- Insert the cell plate into the plate reader. In the instrument panel, select adjust gain, select a random well on the plate, assign it as 5%-10% of the maximum measurable fluorescence value, and select adjust (reading) height.
- Click start measurement to read the whole plate for the background fluorescence signal in relative fluorescence units (RFU).
- "Dual-addition" assay
- Using the liquid handling system with 384/12.5 µL tips, to mix the drug solution in each well of the drug plate, pipette 3x up and down 10 µL of the drug solution (in DPBS containing 10% DMSO) (pipetting speed: 5.2 µL/s), and "aspirate" 1.5 µL from each well of the drug plate (aspiration speed: 1.0 µL/s).
- "Dispense" 0.5 µL of the compounds into the cell assay plate (pipetting speed: 1 µL/s) to reach a final concentration of 2 µM in 0.2% DMSO.
- Place the assay plate immediately into the plate reader after adding the screening compounds. Read the same plate in both the forward and reverse reading directions. To obtain these readouts, define a program to read from "well 1-384" and immediately from "384-1".
NOTE: The readout in both directions takes 2 min in total. This readout design is to compensate for the decrease in signal strength that occurs during plate reading in each direction. See step 3.1 for the readout analyses. - Dispose of the remaining 1 µL of drug solution in the tips by immersing the tips into a 150 mL waste reservoir containing ~50 mL of DPBS (dispensing speed: 1.0 µL/s).
- Incubate the screening compounds with the cells for a total of 5 min (includes the 2 min of plate reading) at room temperature in the biosafety cabinet with the lights off. Add 3 µL of the agonist peptide, Rhimi-K-1 (QFSPWGamide), from the reservoir (aspiration speed: 3.1 µL/s) into the assay plate using the liquid handling system (dispensing speed: 3.1 µL/s) with 384/12.5 µL tips.
- Place the plate into the plate reader immediately after the addition of the agonist peptide. Read the plate in both the forward and reverse directions using the same program as in step 2.3.12.3
3. Data analysis
- Using the analysis software associated with the plate reader installed on a computer, calculate the cellular fluorescence responses (in RFU) for both readings after the first compound addition (First read; RFUago [Supplemental Table S2]) and after the agonist addition steps (Second read = RFUant [Supplemental Table S2]). Each read is obtained by averaging the two values obtained (not shown) by the forward and reverse plate readings from step 2.3.12.3 and step 2.3.12.6, respectively.
NOTE: RFUago, RFUant refer to the relative fluorescence units of the potential agonist hits (read in step 2.3.12.3) and the potential antagonist hits (read in step 2.3.12.6), respectively. - Export all three sets of data, RFUbg, RFUago, and RFUant, from the analysis software into three separate spreadsheets. Each spreadsheet will have only two columns: well position and raw RFU (files not shown here).
NOTE: RFUbg refers to the RFU of the background read in step 2.3.11.2. - Format the data from the three spreadsheets, and organize the above data in one csv file (see the example in Supplemental Table S2). Subtract the background signal read from the RFUago and RFUant, respectively (G and H columns in Supplemental Table S2).
- Import the csv file into a commercially available "online HTS data platform" (see the Table of Materials) for downstream data analysis (Table 1) and storage.
- Manually calculate the Z'-factor for the quality control of each plate assay using equation (1):
equation (1):
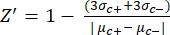 (1)
(1)
NOTE: µc– and µc+ represent the average RFUs of readings of the same wells, which serve as negative controls in the first addition after adding the solvent (blank solvent, n = 64; numbers in blue in Supplemental Table S2) and serve as positive controls after the addition of the agonist (blank solvent + agonist, n = 64; numbers in magenta), respectively. Additionally, σc– and σc+ represent their corresponding standard deviations (SDs). - Manually select the hit molecules from the heat maps on the "online HTS data platform," and calculate the normalized percent activation (NPA) and inhibitory activity (Io) for the agonist hits and antagonist hits using equation (2) and equation (3):
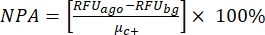 (2)
(2)
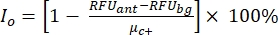 (3)
(3)
Representative Results
An in-house drug plate (SAC2-34-6170) composed of 320 random small molecules was used for demonstrating this HTS assay as an example. The HTS had excellent assay quality with a Z' factor of 0.7 (Table 1). This Z' factor reflects the assay quality independent of the tested compounds34. A Z′-factor of 0.5 or greater indicates a good assay signal dynamic range between the RFUs of the positive controls and the negative controls. Assays with a Z' factor less than 0.5 have a suboptimal signal dynamic range, are usually not informative for hit selection, and are, therefore, discarded. The response (measured in relative fluorescence units, RFU) from positive controls (average RFU of 64 positive controls responses, µc+ = 130,139) was on average 19 times higher than that of the negative controls (average RFU of 64 negative controls responses, µc- = 6,675). The cutoff fluorescence for selecting "agonist hits" here was set to a normalized percent activation (NPA) max ratio of >50%. The cutoff fluorescence for selecting "antagonist hits" was set to an observed inhibitory activity (Io) of >50%. Three agonist hit molecules and two antagonist hits were selected: SACC-0090237 (NPA = 153%), SACC-0036982 (NPA = 118%), and SACC-0006532 (NPA = 152%) as agonists, and SACC-0103392 (Io = 53%) and SACC-0041280 (Io = 56%) as antagonists (the raw data are shown in Supplemental Table S2). Examples of calculations of NPA and Io for agonists (green rows) and antagonists (orange rows), respectively, are shown in Supplemental Table S2.
| Compound addition | µs1 | σs | µc- | σc- | Z’ value |
| (n = 320) | (n = 320) | (n = 64) | (n = 64) | ||
| 9,212 | 18,130 | 6,675 | 1,895 | 0.7 | |
| Agonist peptide addition | µs | σs | µc+ | σc+ | 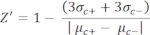 |
| 1,22,322 | 15,987 | 1,30,139 | 11,062 |
Table 1: Summary of the normalized cell responses from the whole plate in the HTS assay. The mean (µ) and standard deviation (σ) are obtained from the CDD vault analysis. Abbreviations: HTS = high-throughput screening; RFUs = relative fluorescence units; 1s (samples) = 320 tested compounds; µs = average value of cellular responses (RFUs) from 320 wells; µc- = average value of RFUs from negative controls; µc+ = average value of RFUs from positive controls; σs = standard deviation of cellular responses (RFUs) from 320 wells; σc- = standard deviation of RFUs from negative controls; σc+ = standard deviation of RFUs from positive controls.
Supplemental Table S1: Custom programs of the liquid handling system for the automatic pipetting steps. This table was modified from Xiong et al.31. Please click here to download this File.
Supplemental Table S2: The raw data of the representative results. The rows highlighted in green represent the agonist hits, and the rows highlighted in orange represent the antagonist hits. Please click here to download this File.
Discussion
The goal of HTS is to identify hit molecules through screening massive numbers of small molecules. Therefore, the results from this example only represent a small part of a conventional HTS experiment. Furthermore, the hit molecules identified need to be validated in downstream assays such as a dose-dependent assay on the same recombinant cell line and on a CHO-K1 cell line carrying only the empty vector, which can be performed simultaneously to save small molecules. Cytotoxicity assays will help demonstrate that a lack of response in the second addition is not due to cell mortality. Other bioassays should be performed to validate activity in the isolated tissues in vitro, or by applying or feeding the small molecules to live arthropods. The complete pipeline of small-molecule ligand identification of the tick kinin receptor can be found in a previous publication31. Through this pipeline, a total of 36 antagonist hits of the tick kinin receptor were identified. Three of these antagonists were further tested in a tissue contraction-inhibition assay and showed anti-myotropic activity on the mosquito hindgut, where orthologous mosquito kinin receptors are expressed31.
Each GPCR-expressing cell line should be validated for suitable experimental conditions before HTS is performed. These conditions include the cell density (10,000 cells per well), the concentration of the screened compounds (2 µM), the final DMSO concentration (0.2%) in the assay, the concentration of the agonist ligand used for the second addition (1 µM), the pipetting speed and depth, and the reading time post the addition of the agonist peptide (5 min). It is critical to adjust the depth and speed of the liquid handling system to ensure that the dispensing steps do not disturb the cells if the cells are adherent. The validation of these conditions before embarking on HTS will aid in troubleshooting if the assay quality is poor. It is important to have the cells in optimal conditions: the cells should reach 90%-100% confluency but not be overconfluent in the cell plate when used for HTS. If the background signals in certain wells are abnormally low or high, after confirming under the microscope for obviously low or high cell density, the outliers should be removed manually when analyzing the data. Before adapting the calcium fluorescence assay to HTS, the kinetic curve of the positive peptide must be known, as the kinetic response of the cells to the agonist peptide should last for 1-2 min for it to be adapted into an HTS assay and last long enough to read the whole plate. To achieve the prolonged signal, a relatively high concentration of the agonist peptide should be applied, or a higher cell density should be tested. If the kinetic response of the certain receptor cells is too short (lasts less than 1 min), it is recommended to read at a faster speed in the end-point readout or use only a portion of the plate for HTS. Certainly, the quality of the recombinant cell line with respect to the level of receptor expression is crucial for success. Different clonal cell lines will likely vary, as the insertion of the plasmid is random in the genome, and this could certainly influence receptor expression. It would be ideal to incorporate an early calcium fluorescence test of the transfected pool and during the process of selecting clonal cell lines.
The limitation of the calcium fluorescence assay is that it measures the activity of the secondary messengers instead of the direct binding of the ligand to the receptor. Although it is possible to exclude the off-target agonist hits by screening the hit molecules on the cells that do not express the target receptor, it is impossible to completely exclude the off-target antagonist hit molecules using this assay because it is possible that some molecules may inhibit the cellular calcium release pathways downstream of the GPCR signaling. Therefore, further validation is needed using either a ligand binding assay or a bioassay for validation at the organism level, such as in a tick tissue assay with a known agonist, as done previously with mosquitos31.
Most functional assays that detect GPCR activation primarily measure intracellular events in two major signaling pathways: G protein-dependent and G protein-independent pathways. G protein-dependent assays measure Ca2+, inositol triphosphate (IP3), or cAMP concentrations, while G protein-independent assays detect primarily receptor trafficking or β-arrestin recruitment7. Different assays utilize different technologies for readouts, such as Ca2+-sensitive dyes (fluorescence assays), photoproteins such as aequorins (Ca2+ bioluminescence assays)14, tagged fluorescence proteins, fluorescence resonance energy transfer (FRET)-based assays, bioluminescence resonance energy transfer (BRET)-based assays, or label-free optical biosensor-based cellular assays. These assays and the corresponding available readout technologies were reviewed in detail by Zhang and Xie35. Some assays are applicable to any GPCRs, and some are specific to certain GPCRs (e.g., the Ca2+ assay for Gq-coupled receptors, cAMP for Gi/o– or Gs– coupled receptors). However, by promiscuously co-expressing certain G proteins with the receptor in the recombinant cells, the signaling pathway can be altered. For example, Gq15/16 proteins can be co-expressed with non-Gq-coupled receptors for PLC-IP3-Ca2+ signaling detection7,36. In contrast to the assays that measure a single molecular event, there are high-content analysis assays, which collect a massive amount of biological data to improve the ligand specificity37. This type of assay is also more complex and expensive to perform7.
In invertebrate research, the most widely used functional assays detect GPCR activation by analyzing the fluctuations in the concentrations of second messengers, such as Ca2+ or cAMP. Due to the differences in the signaling pathways between mammals and invertebrates, most of the other abovementioned readout technologies have not been yet broadly adopted for invertebrate GPCR studies. However, G-proteins are relatively conserved across the animal kingdom. Recently, Lismont et al.38 demonstrated that some human BRET-based G-protein biosensors can also be adapted to detect the activation of an insect GPCR when co-expressed in the receptor recombinant cells. By co-expressing each of the different G protein-based biosensors (Gs, Gi/o, Gq/11, or G12/13) with specific GPCRs, the G protein-coupling mechanism can be elucidated. In the study by Lismont et al.38, the Schgr-CRF-DH could dose-dependently activate the Gi/o and Gq/11 biosensors and did not activate the Gs and G12/13 biosensors.
Overall, the calcium fluorescence assay performed in a 384-well plate in combination with the "dual-addition" approach described here allows the screening of tens of thousands of molecules in a relatively short time. This is a relatively time- and cost-efficient approach to discover hit molecules for downstream validations and applications.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by the USDA-NIFA-AFRI Animal Health and Well-Being Award (Award number 2022-67015-36336, PVP [Project Director]) and from competitive funds from the Texas A&M AgriLife Research Insect Vector Diseases Grant Program (FY'22-23) to P.V.P. The A.W.E.S.O.M.E. faculty group of the College of Agriculture and Life Sciences, TAMU, is acknowledged for help editing the manuscript. Supplemental Table S2 contains data from an in-house, random, small-molecule library obtained from Dr. James Sacchettini's laboratory at Texas A&M University and Texas A&M AgriLife Research.
Materials
| 0.25% trypsin-EDTA | Gibco Invitrogen | 15050-065 | with phenol red |
| 0.4% trypan blue | MilliporeSigma | T8154 | liquid, sterile |
| 1.5 mL microcentrifuge tubes | Thermo Fisher | AM12400 | RNase-free Microfuge Tubes |
| 5 mL serological pipette | Corning | 29443-045 | Corning Costar Stripette individually wrapped |
| 10 mL serological pipette | Corning | 29443-047 | Corning Costar Stripette individually wrapped |
| 15 mL conical tubes | Falcon | 352196 | sterile |
| 20 µL filter tips | USA Scientifc Inc. | P1121 | sterile, barrier |
| 25 mL serological pipette | Corning | 29443-049 | Corning Costar Stripette individually wrapped |
| 50 mL conical tubes | Corning | 430828 | graduated, sterile |
| 150 mL auto-friendly reservior | Integra Bioscience | 6317 | sterile, individually wrapped for cell seeding in day 1 |
| 150 mL auto-friendly reservior | Integra Bioscience | 6318 | sterile, stacked, for loading dye in day 2 |
| 384/ 12.5 µL low retention tips | Integra Bioscience | 6405 | long, sterile filter |
| 384/ 12.5 µL tips | Integra Bioscience | 6404 | long, sterile filter |
| 384-well plate | Greiner | 781091 | CELLSTAR, clear polystyrene, µClear, Black/Flat |
| Aluminum plate seals | Axygen Scientific | PCR-AS-200 | polyester-based |
| Aluminum foil wrap | Walmart | ||
| Biosafty cabinet II | NuAire | NU-540-300 | |
| Cell counter | Nexcelom | AutoT4 | |
| cell counting slides | Nexcelom | SD-100 | 20 µL chamber |
| CO2 humidified incubator | Thermo Fisher | Forma Series II | |
| Desk Lamp | SunvaleeyTEK | RS1000B | |
| Dimethyl sulfoxide | MilliporeSigma | 276855 | anhydous, >99.9% |
| Drug plate | Corning | 3680 | |
| Dulbecco's phosphate-buffered saline | Corning | 21-031-CV | DPBS, 1x without calcium amd magnesium |
| Ethanol | Koptec | 2000 | |
| F-12K Nutrient Mixture | Corning | 45000-354 | (Kaighn's Mod.) with L-glutamine |
| Fetal bovine serum | Equitech-Bio | SFBU30 | |
| Fluorescent calcium assay kit | ENZO Lifescience | ENZ-51017 | 10×96 tests |
| G418 sulfate | Gibco Invitrogen | 10131-027 | Geneticin selective antibiotic 50 mg/mL |
| Hank's buffer | MilliporeSigma | 55037C | HBSS modified, with calcium, with magnesium, without phenol read |
| HEPES buffer | Gibco Invitrogen | 15630-080 | 1 Molar |
| HTS data storage plateform | CDD vault | https://www.collaborativedrug.com/ | |
| Liquid handling system | Integra Bioscience | Viaflo | 384/12.5 µL |
| Plate centrifuge | Thermo Fisher | Sorvall ST8 | |
| Plate reader | BMG technology | Clariostar | |
| Poly-D-lysine | MilliporeSigma | P6407 | |
| Rhimi-K-1 agonist peptide | Genscript | custom order | QFSPWGamide |
| T-75 flask | Falcon | 353136 |
References
- Hanlon, C. D., Andrew, D. J. Outside-in signaling – A brief review of GPCR signaling with a focus on the Drosophila GPCR family. Journal of Cell Science. 128 (19), 3533-3542 (2015).
- Liu, N., Li, T., Wang, Y., Liu, S. G-protein coupled receptors (GPCRs) in insects-A potential target for new insecticide development. Molecules. 26 (10), 2993 (2021).
- Pierce, K. L., Premont, R. T., Lefkowitz, R. J. Seven-transmembrane receptors. Nature Reviews Molecular Cell Biology. 3, 639-650 (2002).
- Pietrantonio, P. V., Xiong, C., Nachman, R. J., Shen, Y. G protein-coupled receptors in arthropod vectors: Omics and pharmacological approaches to elucidate ligand-receptor interactions and novel organismal functions. Current Opinion in Insect Science. 29, 12-20 (2018).
- Hilger, D., Masureel, M., Kobilka, B. K. Structure and dynamics of GPCR signaling complexes. Nature Structural & Molecular Biology. 25 (1), 4-12 (2018).
- Liu, N., Wang, Y., Li, T., Feng, X. G-protein coupled receptors (GPCRs): Signaling pathways, characterization, and functions in insect physiology and toxicology. International Journal of Molecular Sciences. 22 (10), 5260 (2021).
- Hansen, K. B., Bräuner-Osborne, H., Leifert, W. FLIPR® assays of intracellular calcium in GPCR drug discovery. G Protein-Coupled Receptors in Drug Discovery. , (2009).
- Bauknecht, P., Jekely, G. Large-scale combinatorial deorphanization of Platynereis neuropeptide GPCRs. Cell Reports. 12 (4), 684-693 (2015).
- Frooninckx, L., et al. Neuropeptide GPCRs in C. elegans. Frontiers in Endocrinology. 3, 167 (2012).
- Caers, J., et al. More than two decades of research on insect neuropeptide GPCRs: An overview. Frontiers in Endocrinology. 3, 151 (2012).
- Šimo, L., Koči, J., Park, Y. Receptors for the neuropeptides, myoinhibitory peptide and SIFamide, in control of the salivary glands of the blacklegged tick Ixodes scapularis. Insect Biochemistry and Molecular Biology. 43 (4), 376-387 (2013).
- Šimo, L., Park, Y. Neuropeptidergic control of the hindgut in the black-legged tick Ixodes scapularis. International Journal for Parasitology. 44 (11), 819-826 (2014).
- Liesch, J., Bellani, L. L., Vosshall, L. B. Functional and genetic characterization of neuropeptide Y-like receptors in Aedes aegypti. PLoS Neglected Tropical Diseases. 7 (10), 2486 (2013).
- Lu, H. L., Kersch, C. N., Taneja-Bageshwar, S., Pietrantonio, P. V. A calcium bioluminescence assay for functional analysis of mosquito (Aedes aegypti) and tick (Rhipicephalus microplus) G protein-coupled receptors. Journal of Visualized Experiments. (50), e2732 (2011).
- Xiong, C., Baker, D., Pietrantonio, P. V. The cattle fever tick, Rhipicephalus microplus, as a model for forward pharmacology to elucidate kinin GPCR function in the Acari. Frontiers in Physiology. 10, 1008 (2019).
- Holmes, S. P., Barhoumi, R., Nachman, R. J., Pietrantonio, P. V. Functional analysis of a G protein-coupled receptor from the Southern cattle tick Boophilus microplus (Acari: Ixodidae) identifies it as the first arthropod myokinin receptor. Insect Molecular Biology. 12 (1), 27-38 (2003).
- Cox, K. J., et al. Cloning, characterization, and expression of a G-protein-coupled receptor from Lymnaea stagnalis and identification of a leucokinin-like peptide, PSFHSWSamide, as its endogenous ligand. Journal of Neuroscience. 17 (4), 1197-1205 (1997).
- Dircksen, H., Kastin, A. J. Chapter 32 – Crustacean bioactive peptides. Handbook of Biologically Active Peptides (Second Edition). , 209-221 (2013).
- Halberg, K. A., Terhzaz, S., Cabrero, P., Davies, S. A., Dow, J. A. Tracing the evolutionary origins of insect renal function. Nature Communications. 6, 6800 (2015).
- Pietrantonio, P. V., Jagge, C., Taneja-Bageshwar, S., Nachman, R. J., Barhoumi, R. The mosquito Aedes aegypti (L.) leucokinin receptor is a multiligand receptor for the three Aedes kinins. Insect Molecular Biology. 14 (1), 55-67 (2005).
- Radford, J. C., Davies, S. A., Dow, J. A. Systematic G-protein-coupled receptor analysis in Drosophila melanogaster identifies a leucokinin receptor with novel roles. Journal of Biological Chemistry. 277 (41), 38810-38817 (2002).
- Brock, C. M., et al. The leucokinin-like peptide receptor from the cattle fever tick, Rhipicephalus microplus, is localized in the midgut periphery and receptor silencing with validated double-stranded RNAs causes a reproductive fitness cost. International Journal for Parasitology. 49 (3-4), 287-299 (2019).
- Nässel, D. R. Leucokinin and associated neuropeptides regulate multiple aspects of physiology and behavior in Drosophila. International Journal of Molecular Sciences. 22 (4), 1940 (2021).
- Kim, Y. -. J., et al. Central peptidergic ensembles associated with organization of an innate behavior. Proceedings of the National Academy of Sciences of the United States of America. 103 (38), 14211-14216 (2006).
- Al-Anzi, B., et al. The leucokinin pathway and its neurons regulate meal size in Drosophila. Current Biology. 20 (11), 969-978 (2010).
- Yurgel, M. E., et al. A single pair of leucokinin neurons are modulated by feeding state and regulate sleep-metabolism interactions. PLoS Biology. 17 (2), 2006409 (2019).
- Nässel, D. R., Zandawala, M. Recent advances in neuropeptide signaling in Drosophila, from genes to physiology and behavior. Progress in Neurobiology. 179, 101607 (2019).
- Okusawa, S., Kohsaka, H., Nose, A. Serotonin and downstream leucokinin neurons modulate larval turning behavior in Drosophila. Journal of Neuroscience. 34 (7), 2544-2558 (2014).
- Kersch, C. N., Pietrantonio, P. V. Mosquito Aedes aegypti (L.) leucokinin receptor is critical for in vivo fluid excretion post blood feeding. FEBS letters. 585 (22), 3507-3512 (2011).
- Kwon, H., et al. Leucokinin mimetic elicits aversive behavior in mosquito Aedes aegypti (L.) and inhibits the sugar taste neuron. Proceedings of the National Academy of Sciences of the United States of America. 113 (25), 6880-6885 (2016).
- Xiong, C., Baker, D., Pietrantonio, P. V. A random small molecule library screen identifies novel antagonists of the kinin receptor from the cattle fever tick, Rhipicephalus microplus (Acari: Ixodidae). Pest Management Science. 77 (5), 2238-2251 (2021).
- Torfs, P., et al. The kinin peptide family in invertebrates. Annals of the New York Academy of Sciences. 897 (1), 361-373 (1999).
- Ma, Q., Ye, L., Liu, H., Shi, Y., Zhou, N. An overview of Ca2+ mobilization assays in GPCR drug discovery. Expert Opinion on Drug Discovery. 12 (5), 511-523 (2017).
- Zhang, J. -. H., Chung, T. D., Oldenburg, K. R. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. Journal of Biomolecular Screening. 4 (2), 67-73 (1999).
- Zhang, R., Xie, X. Tools for GPCR drug discovery. Acta Pharmacologica Sinica. 33 (3), 372-384 (2012).
- Offermanns, S., Simon, M. I. Gα15 and Gα16 couple a wide variety of receptors to phospholipase C. Journal of Biological Chemistry. 270 (25), 15175-15180 (1995).
- Murgia, M. V., et al. High-content phenotypic screening identifies novel chemistries that disrupt mosquito activity and development. Pesticide Biochemistry and Physiology. 182, 105037 (2022).
- Lismont, E., et al. Can BRET-based biosensors be used to characterize G-protein mediated signaling pathways of an insect GPCR, the Schistocerca gregaria CRF-related diuretic hormone receptor. Insect Biochemistry and Molecular Biology. 122, 103392 (2020).

