Pancreatic Tissue Dissection to Isolate Viable Single Cells
Summary
Pancreatic metaplastic cells are precursors of malignant cells that give rise to pancreatic tumors. However, isolating intact viable pancreatic cells is challenging. Here, we present an efficient method for pancreatic tissue dissociation. The cells can then be used for single-cell RNA sequencing (scRNA-seq) or for two- or three-dimensional co-culturing.
Abstract
The pancreas includes two major systems: the endocrine system, which produces and secretes hormones, and the exocrine system, which accounts for approximately 90% of the pancreas and includes cells that produce and secrete digestive enzymes. The digestive enzymes are produced in the pancreatic acinar cells, stored in vesicles called zymogens, and are then released into the duodenum via the pancreatic duct to initiate metabolic processes. The enzymes produced by the acinar cells can kill cells or degrade cell-free RNA. In addition, acinar cells are fragile, and common dissociation protocols result in a large number of dead cells and cell-free proteases and RNases. Therefore, one of the biggest challenges in pancreatic tissue digestion is recovering intact and viable cells, especially acinar cells. The protocol presented in this article shows a two-step method that we developed to meet this need. The protocol can be used to digest normal pancreata, pancreata that include pre-malignant lesions, or pancreatic tumors that include a large number of stromal and immune cells.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the most aggressive cancer types1. Clinical evidence supports the notion that PDAC develops from exocrine-system cells, including acinar cells, over many years, driven by mutations in the KRAS proto-oncogene2.
Pancreatic tumors include many different cell types, and it has been demonstrated that malignant cells count for only 20%-50% of the tumor mass3. Different cell types interact with the epithelial cells, support their transformation, and enhance tumor formation and growth. Early events cause acinar metaplasia, which gives rise to microscopic lesions called pancreatic intraepithelial neoplasia (PanINs), which can in some cases develop into PDAC4.
There is a critical need to investigate these interactions and target pivotal signals. Single-cell RNA-sequencing (scRNA-seq) is a powerful method that reveals gene expression at a single-cell resolution, thereby tracking the changes that epithelial cells undergo, thus enabling the exploration of pancreatic cancer development.
Tissue dissection and digestion to single cells is the first stage in a scRNA-seq experiment. Several factors make pancreatic tissue digestion especially challenging: i) acinar cells account for more than 90% of the pancreas and acinar cells contain large amounts of digestive enzymes, including proteases and RNases that reduce the quality of RNA-based libraries; (ii) acinar cells are very sensitive and may lyse if standard protocols are used; (iii) acinar cells express a small number of genes at very high levels. Therefore, if these cells are lysed during the experiment, this can contaminate the observed gene expression profile of other cells; (iv) pancreatic tissue recovered from tumors is desmoplastic, making it hard to dissect without damaging the cells. Thus, even though maintaining high viability of all the cell types is required, the large number and sensitivity of acinar cells add additional complexity. These factors impose difficulties in achieving a single-cell suspension that is more than 80% viable and has no clumps, as is required for scRNA-seq experiments.
Here, we developed a protocol using trypsin C and collagenase P, along with frequent tissue monitoring. This supports dissociation to single cells while retaining high viability to support the success of scRNA-seq experiments5,6.
Protocol
The joint ethics committee (Institutional Animal Care and Use Committee) of the Hebrew University (Jerusalem, Israel) and Hadassah Medical Center (Jerusalem, Israel) approved the study protocol for animal welfare (MD-18-15417-5 "Tissue dynamics in pancreatic cancer in mice"), and the protocol presented here complied with all relevant ethical regulations for animal testing and research. The Hebrew University is an Association for Assessment and Accreditation of Laboratory Animal Care International-accredited institute.
NOTE: The mouse strain stock #007908, stock #019378, and stock #008179 were obtained from Jackson's laboratory. PRT (Kras+/LSL-G12D; Ptf1a-CreER; Rosa26LSL-tdTomato) mice were created by crossing the above strains. Mice from both genders, between 6 weeks and 15 months of age, were used for the study. Tamoxifen was prepared by dissolving the powder in corn oil. Adult mice (6-8 weeks of age, females and males), were injected with tamoxifen subcutaneously on days 0 and 2 at a dose of 400 mg/kg and examined twice a week following the injection. It was not possible to measure tumors as they were internally located; therefore, euthanasia was performed if abnormal clinical signs were observed according to the ethical protocol. Mice were euthanized at different time points post-tamoxifen induction, using isoflurane and cervical dislocation.
1. Pancreatic dissection
NOTE: For optimal yield during extraction and to ensure good cell viability, rapid dissection is critical. To shorten the time required for pancreas isolation, all instruments and equipment must be ready on ice before euthanizing the mouse.
- Euthanize the mouse by CO2 asphyxiation and verify using cervical dislocation. From this step on, all procedures must be performed with sterile dissecting instruments.
- Fix the mouse and spray the abdomen with 70% ethanol. Make a V-shaped incision of 2.5 cm in the genital area with scissors and forceps and proceed upward to fully open the abdominal cavity.
- Locate the stomach on the left side of the mouse. Locate the pancreas, which is near the spleen. Separate the pancreas from the stomach and duodenum using two forceps (without tearing). Continue and separate the pancreas from the small intestine, jejunum, and ileum.
- Move the pancreas to the right side of the mouse. Separate the remaining connections between the pancreas and the thoracic cavity with forceps to completely detach the pancreas and attached spleen.
- Remove the pancreas and spread it out for examination in a Petri dish on ice.
NOTE: Care must be taken during this step to remove only the pancreas, and not remove mesenteric fat tissue or other adjacent tissue along with the pancreas, to avoid cellular contamination.
2. Enzymatic and mechanical dissociation of the pancreas
- Prepare the following buffers in advance.
- Dissociation buffer 1: 4 mL of trypsin C + 6 mL of phosphate buffered saline (PBS) (see Table 1) for each sample.
- Dissociation buffer 2: 9 mL of Hanks′ balanced salt solution (HBSS) 1x; 4% bovine serum albumin (BSA); 1 mL of collagenase P (10 mg/mL); 200 µL of trypsin inhibitor (10 mg/mL); and 200 µL of DNase I (10 mg/mL) (see Table 1).
- Wash buffer: 50 mL of HBSS 1x; 2 g of BSA; 1 mL of trypsin inhibitor (10 mg/mL); and 1 mL of DNase 1 (10 mg/mL).
- Enzyme activity stop solution: HBSS 1x containing 5% fetal bovine serum (FBS) and 150 g of DNase I (0.2 mg/mL).
- Place the pancreas in a 50 mL tube on ice. Rinse the pancreas in 10% FBS in HBSS 1x. The fatty tissue will float and the pancreas will sink. This is an easy way to visualize and quickly remove the contaminating white adipose tissue still attached to the pancreas.
- Transfer the mouse pancreatic tissue to a sterile Petri dish containing 5 mL of HBSS 1x on ice. Cut the pancreas into small pieces of 1 to 3 mm3 using Noyes scissors and a scalpel (Figure 2A). In case of more than one sample, the samples should be kept on ice in 10% FBS/HBSS 1x.
- Transfer the tissues to a centrifuge tube. Centrifuge at 350 x g at 4 °C for 5 min. Aspirate and discard the supernatant to remove cell fragments and blood cells.
- Resuspend the pieces in dissociation buffer 1 containing 0.02% trypsin C-0.05% EDTA for 10 min at 37 °C with agitation (180 rpm). Immediately wash with 10% FBS/Dulbecco's modified Eagle's medium (DMEM). Centrifuge for 5 min at 350 g at 4 °C.
- Wash again by resuspending the pellet in 10 mL of wash buffer and centrifuging at 350 x g for 5 min at 4 °C before the next dissociation step.
- Incubate the pancreas in dissociation buffer 2 for 15 min at 37 °C with agitation (180 rpm).
- After 15 min, perform mechanical dissociation by vigorously pipetting the pancreatic fragments up and down in sterile pipettes of decreasing size (25, 10, and 5 mL serological pipettes) 10 times and bring back to 37 °C.
- After an additional 5 min, repeat the mechanical dissociation and use light microscopy to monitor the dissociation according to the amount of single-cell suspension. Usually if at this stage, less than 90% of the suspension consists of isolated single cells, a longer incubation time is needed. Use trypan blue to monitor cell viability.
- Continue with the incubation and take samples to detect the dissociation every 5 min.
NOTE: The total incubation time depends on the tissue and can vary between samples. The incubation and tissue dissociation should end when 90% of the cells are separated into single cells or when a reduction in viability is detected.
- After the pancreatic tissue is well dissociated (corresponding to the disappearance of pancreatic fragments and the increasing turbidity of the solution) (Figure 2), stop the enzymatic reaction by washing twice with enzyme activity stop solution for 5 min at 4 °C. From this step, keep the cell suspension on ice.
- Pass the cell suspension through a 70µm nylon mesh and check the cell viability under the microscope. Smaller sizes of nylon mesh may reduce cell viability.
- Resuspend and wash the pellet with 5-10 mL of ice-cold buffered wash solution. Count the cells.
- If several red blood cells are observed, treat the cells with a red blood cell lysis buffer for 2 min at room temperature. In cases where clumps are observed, the sample should be treated again with trypsin, as described in step 2.5. Viability is detected with trypan blue under the microscope, and if viability is less than 80%, live cells should be isolated using the magnetic-activated cell sorting (MACS) dead cells removal kit with MACS MS columns (see Table 1).
Representative Results
In a recently published work5, we applied the protocol described above to explore the early stages of PDAC development using a mouse model. The mouse was genetically engineered to include the cassettes Ptf1a–CreER, LSL-Kras-G12D, LSL-tdTomato7, which allow the expression of constitutively active KRAS in acinar cells after tamoxifen injection.
After cervical dislocation (according to the mouse ethical protocol), the pancreas was resected, and the protocol described above was applied (see Figure 1 and Protocol).
To optimize the dissociation protocol, the dissociation was performed with or without pre-incubation of the tissue with trypsin C. It was found that pre-incubation with trypsin C accelerated the dissociation without affecting the viability. In addition, different collagenases, including collagenase D, collagenase 1a, and collagenase P, were tried (see Table 1). It was found that using collagenase D resulted in massive cell death, and indeed in other studies that used this collagenase, the fraction of acinar cells was very small8. The use of collagenase 1a also did not provide the expected results, as even after a 90 min incubation with collagenase 1a, the tissue was not dissociated. Only collagenase P allowed us to dissociate the tissue and maintain high viability of all the cell types.
These experiments were repeated at seven different time points post-tamoxifen injection. Simultaneously with acinar-to-ductal metaplasia and PanIN lesion formation, there was an accumulation of stromal and immune cells, including fibroblasts, and the tissue became desmoplastic and stiff. The different time points post-tamoxifen injection allowed us to examine the protocol under several different tissue states and measure the recovery of epithelial, stromal, and immune cells.
A photo of a mouse is shown in Figure 2A and the isolated pancreas, a fluffy tissue, is shown in Figure 2C. The red color of the tissue results from the expression of tdTomato in the acinar cells. The protocol was used successfully with all the tissue states and with human samples. However, incubation times with trypsin and collagenase may vary. It is therefore important to monitor the sample and observe the dissociation of the tissue and cell viability every few minutes under the microscope. At an early stage of the dissociation protocol, there were low numbers of isolated cells and large numbers of clumps (Figure 2D, left). Our aim was to achieve isolated viable cells, as shown in Figure 2D, middle, and avoid a reduction in the number of viable cells (Figure 2D, right).
It is important to note that longer incubation times reduced the viability of the cells. Starting from a tissue 0.5 x 10 cm3 in size, we recovered a total of 5 x 106 cells, 5 x 105 of which were tdTomato positive cells. According to the fluorescence-activated cell sorting (FACS) analysis in Figure 3, our pancreas dissociation protocol supports the recovery of multiple cell types, including acinar cells, ductal cells, endothelial cells, fibroblasts, immune cells, and pericytes with high viability. However, the actual ratio between the number of cells from each type in the tissue before dissociation may be different, inferred from the fluorescence images of pancreatic frozen sections that include tdTomato positive cells (Figure 2E). The high viability achieved using the protocol detailed above is shown by the FACS analysis (Figure 3) and the live cell/dead cell counting done using a hemocytometer (Figure 3H).
For each sample, we performed scRNA-seq and, consistent with the FACS analysis, we confirmed the detection of all the above-mentioned cell types (Figure 4A,B), in each of the resected tissue from all the time points post-tamoxifen injection. We also examined the expression of carboxypeptidase 1 (Cpa1), which encodes carboxypeptidase1. Cpa1 expression in acinar cells is extremely high and can indicate to what extent acinar cells were lysed during the dissociation, as well as the extent to which they have contaminated the transcriptome of other cell types. We detected minimal contamination, as can be seen in Figure 4C,D. The gentle dissociation also results in high viability that allows us to follow-up with cell sorting of desired cell types for additional experiments (Figure 3).
Together, the quality of the dissociation protocol supports different follow-up experiments, including scRNA-seq.
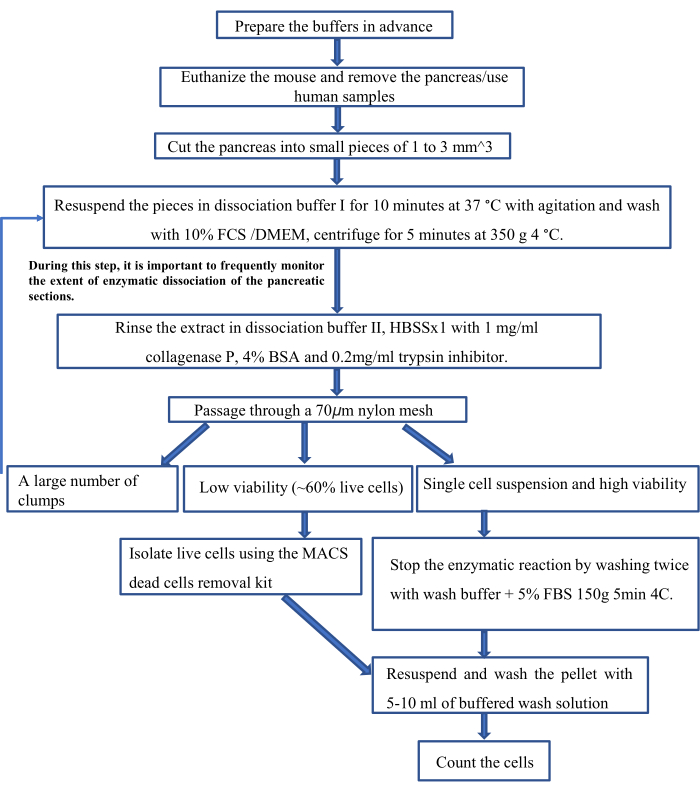
Figure 1: Scheme of the protocol. Please click here to view a larger version of this figure.
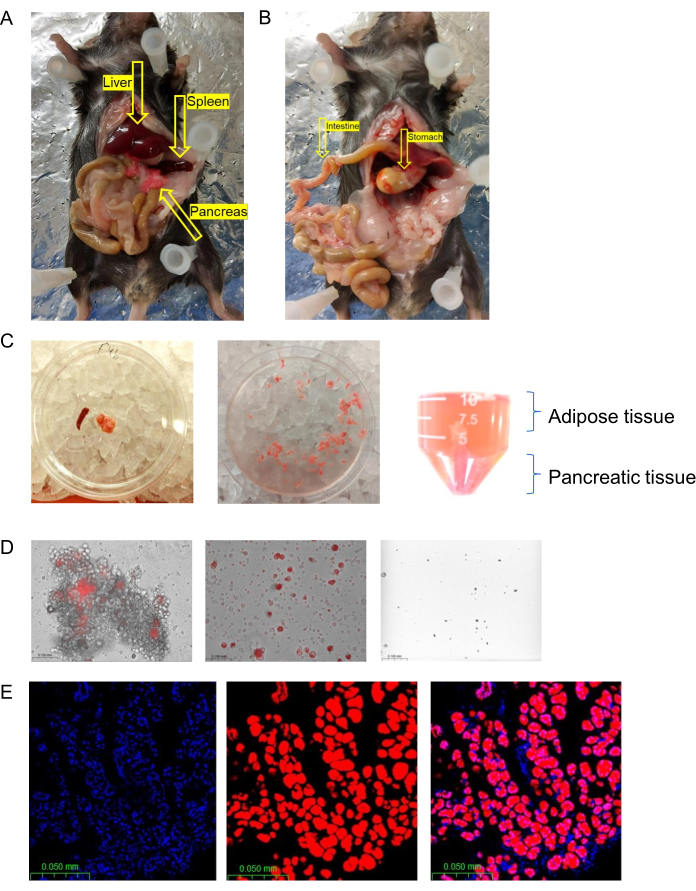
Figure 2: Pancreatic cell isolation. (A) A photo showing the pancreas (in red), liver, and spleen of the mouse after fully opening the abdominal cavity. (B) The same mouse as in (A) after removal of the pancreas. (C) The dissection of the pancreas. The pancreas after removal from the animal (left). The pancreas after cleavages into small pieces (middle). Phase separation after centrifugation (right). (D) Monitoring the cells under the microscope to examine single-cell viability. Total primary pancreatic cell isolation (20x). Clumps of cells after 15 min of enzymatic reaction (left). Single-cell suspension after 25 min of enzymatic reaction (middle). Reduced cell viability after a long incubation (right). (E) Fluorescence images of a frozen pancreatic section. Acinar cells are tdTomato positive; DAPI staining in blue (photo at 40x). Please click here to view a larger version of this figure.
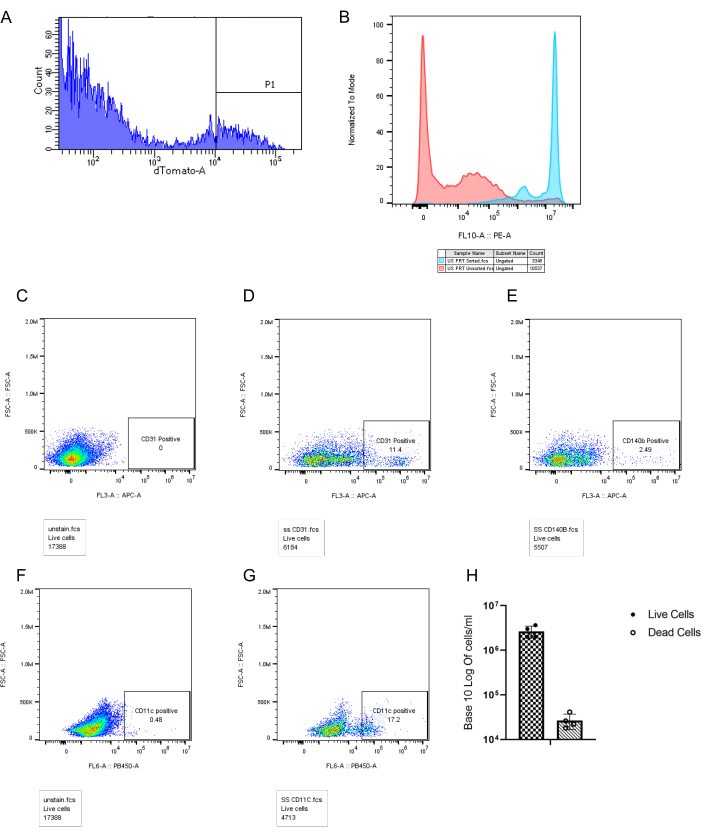
Figure 3: FACS analysis of single-cell suspension of pancreatic cancer. (A) tdTomato positive and negative cells. (B) Flow cytometry of unsorted (red) versus sorted (blue) acinar cells. (C,F) Unstained cells, APC and PB450. (D) FACS analysis after staining with Anti-CD31, a marker of endothelial cells. (E) FACS analysis after staining with Anti-CD140b, a marker of pericytes. (G) FACS analysis after staining with Anti-CD11c, a marker of dendritic cells and macrophages. (H) Ratio of live cells and dead cells in four different pancreas cell isolations. Please click here to view a larger version of this figure.
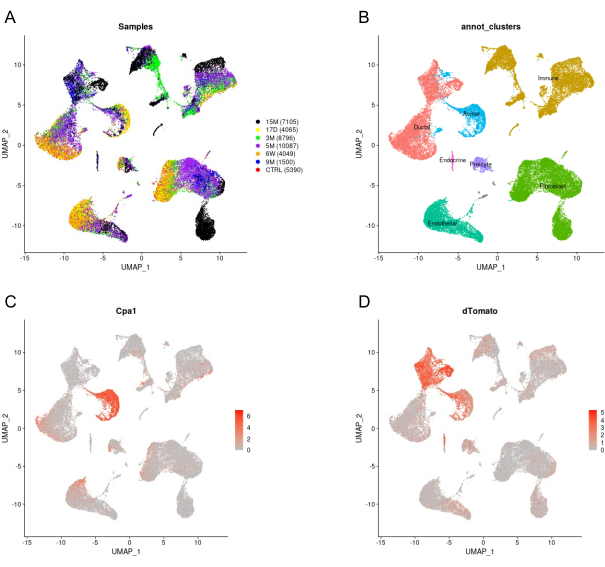
Figure 4: Analysis of scRNA-seq data that was produced using the protocol that we describe in the current article. (A) UMAP showing scRNA-seq of a mouse pancreas at different time points post-tamoxifen injection, as indicated in the right side of the panel. (B) Cell types were determined based on known markers. (C,D) Cells are colored according to the expression level of Cpa1 (in C) or the level of tdTomato (in D). Please click here to view a larger version of this figure.
Discussion
In this article, we present a protocol for pancreatic tissue dissociation. The protocol is simple, easy to use, and provides a tool to isolate viable single cells from pancreatic tissue at different stages during the malignancy process, including solid tumors. In previous studies, different types of collagenases were used to digest the pancreas8,9. Using a very potent collagenase, such as collagenase D, results in a large population of immune cells and a lower percentage of epithelial cells. The use of collagenase P allows pancreatic digestion that is suitable for intact or tumor pancreatic tissue.
Based on our observations, all the cell types can be isolated. We previously validated the infiltration of immune T cells, for example, using immunohistochemistry5; we believe that the relative cell number that we observed in our analysis is a proper approximation to the actual representation of cells in the tissue, except for acinar cells that are fragile and were therefore underrepresented. We recommend using immunohistochemistry or spatial transcriptomics10,11,12 if there is a need for accurate proportions of cell types in the tissue. In addition, we observed that the dissociation of tissue from wild-type mice in the absence of constitutively active KRAS expression is more challenging and requires more frequent monitoring, to stop the enzymatic reaction on time by adding 10% FBS to the wash buffer and avoid massive cell death. This is also consistent with contamination from some highly abundant acinar transcripts, such as Cpa1, although our protocol minimizes this problem.
The cell isolation can be used for scRNA-seq, as we showed5, for culturing cells, or as a starting material for organoids, for example.
The need for fresh tissue is one limitation of the method. An alternative approach that focuses on the isolation of nuclei, single-nucleus RNA sequencing (sNuc-seq)13, was recently used to investigate PDAC tumors from patients at a single-cell resolution11; in the future, it will be interesting to compare the data quality of pancreatic epithelial cells that are obtained from sNuc-seq compared to scRNA-seq. Importantly, nucleus extraction does not allow for cell culturing, and sorting to enrich for specific cell types is extremely challenging. Therefore, the tissue dissociation protocol will be useful in the future for scRNA-seq experiments and for these additional applications.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We would like to thank Dr. Avital Sarusi-Portuguez for help in data analysis and Dr. Dror Kolodkin-Gal for assistance in establishing the protocol in a previous study. We thank all past and present members of the Parnas lab. We thank Dr. Gillian Kay and Dr. Michael Kanovsky for their help in editing. This project has received funding from the Israel Science Foundation grant (No. 526/18 O.P.), the Alex U. Soyka Program, and a grant from the Israel Cancer Research Fund (Research Career Development Award).
Materials
| Reagent or Resource | |||
| 70 µm nylon mesh | Corning | cat##431751 | |
| BSA | Sigma Aldrich | cat# A7906 | |
| Collagenase P | Roche | cat# 11213857001 | |
| Critical Commercial Assay | |||
| DAPI | Sigma Aldrich | cat#MBD0015 | |
| Dnase I | Roche | cat# 10104159001 | |
| Experimental Models: Organisms/Strains | |||
| Fetal Bovine Serum South American | ThermoFisher | Cat#10270106 | |
| Hanks' Balanced Salt Solution | Biological industries | cat#02-018-1A | |
| KRASLSL-G12D mice | Jackson Laboratory | JAX008179 | |
| MACS dead cells removal kit | Milteny Biotec | cat#130-090-101 | |
| PBS | Biological industries | cat#02-023-1A | |
| Ptf1a-CreER mice | Jackson Laboratory | JAX019378 | |
| Ptf1a-CreER; Rosa26LSL-tdTomato mice | Jackson Laboratory | JAX007908 | |
| Trypsin C-EDTA 0.05% | Biological industries | cat# 03-053-1A | |
| Trypsin inhibitor | Roche | cat#T6522 |
References
- Siegel, R. L., Miller, K. D., Jemal, A. Cancer statistics, 2019. CA: a Cancer Journal for Clinicians. 69 (1), 7-34 (2019).
- Yachida, S., et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 467 (7319), 1114-1117 (2010).
- Peng, J., et al. Author correction: single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Research. 29 (9), 777 (2019).
- Hruban, R. H., Wilentz, R. E., Kern, S. E. Genetic progression in the pancreatic ducts. The American Journal of Pathology. 156 (6), 1821-1825 (2000).
- Schlesinger, Y., et al. Single-cell transcriptomes of pancreatic preinvasive lesions and cancer reveal acinar metaplastic cells’ heterogeneity. Nature Communications. 11 (1), 4516 (2020).
- Kolodkin-Gal, D., et al. Senolytic elimination of Cox2-expressing senescent cells inhibits the growth of premalignant pancreatic lesions. Gut. 71 (2), 345-355 (2021).
- Kopp, J. L., et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 22 (6), 737-750 (2012).
- Elyada, E., et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discovery. 9 (8), 1102-1123 (2019).
- Bernard, V., et al. Single-cell transcriptomics of pancreatic cancer precursors demonstrates epithelial and microenvironmental heterogeneity as an early event in neoplastic progression. Clinical Cancer Research. 25 (7), 2194-2205 (2019).
- Moncada, R., et al. Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nature Biotechnology. 38 (3), 333-342 (2020).
- Hwang, W. L., et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nature Genetics. 54 (8), 1178-1191 (2022).
- Cui Zhou, D., et al. Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Nature Genetics. 54 (9), 1390-1405 (2022).
- Habib, N., et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nature Methods. 14 (10), 955-958 (2017).

