Isolation, Processing and Analysis of Murine Gingival Cells
Summary
This study describes an efficient technique to isolate and process gingival tissues from the mouse oral cavity in order to produce a single-cell culture. The resulting cells can be further used for flow cytometry analysis and molecular studies.
Abstract
We have developed a technique to precisely isolate and process murine gingival tissue for flow cytometry and molecular studies. The gingiva is a unique and important tissue to study immune mechanisms because it is involved in host immune response against oral biofilm that might cause periodontal diseases. Furthermore, the close proximity of the gingiva to alveolar bone tissue enables also studying bone remodeling under inflammatory conditions. Our method yields large amount of immune cells that allows analysis of even rare cell populations such as Langerhans cells and T regulatory cells as we demonstrated previously 1. Employing mice to study local immune responses involved in alveolar bone loss during periodontal diseases is advantageous because of the availability of various immunological and experimental tools. Nevertheless, due to their small size and the relatively inconvenient access to the murine gingiva, many studies avoided examination of this critical tissue. The method described in this work could facilitate gingival analysis, which hopefully will increase our understating on the oral immune system and its role during periodontal diseases.
Introduction
Gingiva is the soft tissue surrounding the cervical portion of the teeth and covers the alveolar process (Figure 1). The gingiva is a type of masticatory mucosa that can be further separated into mucosal epithelium and connective tissue (also known as submucosa or lamina propria). The anatomical structure of the gingiva and adjacent teeth allows bacteria to reside and develop plaque (biofilm) that constantly challenges the local immune system. As a result, inflammatory response develops in the gingiva, which in certain circumstances becomes destructive – a condition termed periodontal diseases 2. Basically, plaque-induced periodontal pathologies can be divided into gingivitis and periodontitis. Gingivitis represents a reversible condition of local inflammatory response that is confined to the gingiva. Periodontitis, on the other hand, is an irreversible destructive process in which the attachment apparatus (alveolar bone, periodontal ligament, cementum and gingiva) is destroyed 3.
The gingiva was proposed to serve both as effector and inductive sites during periodontal diseases 4. Human studies have suggested that in response to dental plaque, immune effector cells and molecules dynamically infiltrated or departure the gingiva 5-7. This activity was shown to play a major role in periodontal destruction 8,9. Whereas data generated by those studies provided valuable information regarding this pathological process, working with human tissues possess major ethical, technical and experimental limitations. The development of experimental models allowed cause-effect experimentations via employing transgenic mice and in vivo interventions 10. As a result, our knowledge on the mechanisms involved in periodontal disease increased considerably during the last two decades. Even so, due to the complexity of periodontal diseases, there is an ongoing debate regarding the nature of local immune response facilitating tissue destruction. There is also a lack in our understanding on the function of central immune cells in the gingiva during periodontal diseases. It is thus essential to study pathological inflammatory events occurring in the target tissue of the disease, the gingiva.
Protocol
Prepare in advance:
- PBS + 2% FCS
- PBS + 2% FCS with 2 mg/ml of Collagenase Type II and 1 mg/ml of DNAse Type I (1 ml per sample)
- Sterilized surgical instruments
- 0.5 M EDTA solution
1. Upper Gingival Excision Technique
- Euthanize mice using approved IACUC guidance.
- Cut both sides of the oral cavity including the cheeks and the mandible ramus with a sharp/blunt straight scissors.
- Pull down the mandible.
- Cut the soft and hard tissues in an imaginary line 1 mm behind the thirds molars with standard scissors. Direct the scissors perpendicular to the plane of the palatal bone (Figure 2A, blue lines).
- Incise the soft and hard tissues 2 mm behind the incisors (Figure 2A, blue lines).
- Pull down the anterior part of the mouth. The nasal cavity is observable after this step.
- Place the scissors parallel to the palate, put one blade into the nasal cavity, the other blade should incise at the vestibule. Cut until the posterior incision (Figure 2A, green lines). Repeat this step on both sides of the maxilla. At the end the maxilla is detached from the rest of the skull (Figure 2B).
- Remove the maxilla, cut it in the middle suture (Figure 2, black dashed line) and trim the palatal tissue of each hemi-maxilla until reaching the alveolar bone.
- Peel the gingival tissues (both hemi-maxillae) from their anterior border with Adson forceps (without teeth).
- Place the excised gingiva in a plate with PBS + 2% FCS.
2. Gingival Processing
- Put the excised gingiva in a tissue culture dish (35 x 10 mm) with 1 ml PBS + 2% FCS, 2 mg/ml of Collagenase Type II and 1 mg/ml of DNAse Type I.
- Mince well the tissue with an N°15 sterile surgical blade.
- Transfer the tissue from the plate to a 15 ml conical test tube.
- Wash the tissue/cells remaining on the plate with 1 ml PBS + 2% FCS and transfer to the same test tube.
- Incubate in a shaker incubator for 20 min at 37 °C, 200 rpm (recommendation: preheat the incubator).
- Add 20 μl of EDTA 0.5 M and incubate for another 10 min at 37 °C, 200 rpm.
- Fill the test tube up to 12 ml with PBS + 2% FCS and centrifuge at 4 °C, 400 x g for 8 min (recommendation: pre-chill the centrifuge).
- Remove supernatant and resuspend cells with 2 ml PBS + 2% FCS.
- Filter the sample with a 70 μm cell-strainer and save flow through.
- Centrifuge at 4 °C, 320 x g for 5 min.
- Remove supernatant and resuspend cells with 300 μl PBS + 2% FCS.
- Count the cells (approximately 5-10 x 105 cells are expected from a single maxilla).
3. Extracellular and Intracellular Antibody Staining for Flow Cytometry Analysis
It is recommended to work inside a hood, with lights off. It is critical to keep the cells in constant cold environment, as well as all the materials needed.
- Stain cells against extracellular molecules by adding 0.2-0.5 μg of each selected antibody per 1 x 106 cells in a total volume of 100 μl.
- Vortex briefly.
- Incubate the tubes for 15 min in the dark at 4 °C.
- Wash cells with 2 ml PBS + 2% FCS.
- Centrifuge at 4 °C, 320 x g for 5 min.
- Aspirate supernatant and resuspend cells by adding, drop wise, 500 μl cold BD Cytoperm/Cytofix while vortexing.
- Incubate the tubes for 40 min at 4 °C in the dark.
- Vortex briefly.
- Wash the cells twice with 1 ml cold BD Perm/Wash Buffer and spin at 800 x g for 7 min at 4 °C.
- Aspirate supernatant except for 300 μl.
- Stain cells intracellularly by adding 1 μg of each selected antibody per 1 x 106 cells in a total volume of 100 μl.
- Vortex briefly.
- Incubate for 30 min in the dark at 4 °C.
- Wash the cells twice with 1 ml cold BD Perm/Wash Buffer and spin at 800 x g for 7 min at 4 °C.
- Aspirate supernatant and resuspend cells by adding 300 μl cold BD Perm/Wash Buffer.
- Vortex briefly.
- Transfer the suspension while filtering to FACS tubes.
- Keep the tubes in a dark and cold environment until analysis.
Representative Results
Examples of flow cytometry analysis on gingival cells are presented. Gingival cells pooled from 2 mice were run in an LSR II flow cytometer and analyzed using FlowJo software. Figure 3A demonstrated the distribution of gingival cells from naïve mice in a side scatter (SSC) versus forward scatter (FSC) plot. Gating strategy to identify (i) lymphocytes (ii) monocytes / dendritic cells and (iii) granulocytes is indicated. For comparison purpose, we also present a FACS plot illustrating gingival cells purified from P. gingivalis infected mice 21 days after oral gavage in a setting of experimental periodontitis as we previously described (Figure 3B). An increase in the various immune cell populations can be easily detected in the infected mice as compared to naïve mice. Next, we identify immune cells in the processed gingiva by gating on cells expressing the hematopoietic marker CD45 (Figure 4A). These cells further segregated into CD3-positive cells representing T cells (Figure 4B). We also identified gingival neutrophils according to expression of the Ly6G and CD11b molecules (Figure 4C). Finally, langerhans cells, an epithelial subset of dendritic cells (DCs), were identified by gating on MHC class II+CD11c+ cells (a marker of murine DCs) and expression of Ep-CAM and langerin/CD207 (intracellular staining) (Figure 4D).
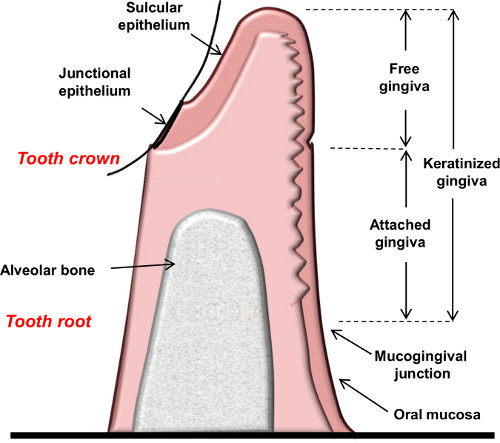
Figure 1. Schematic diagram showing the anatomical landmarks of the gingiva.
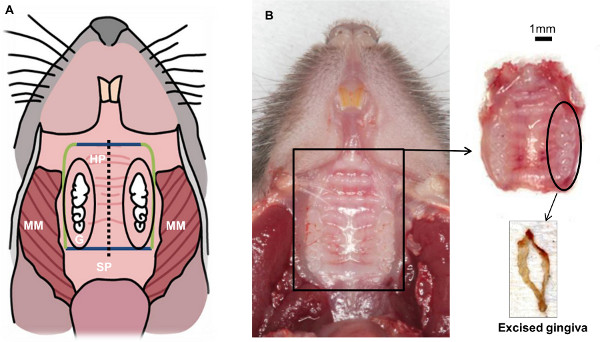
Figure 2. Schematic view of mouse oral cavity resulting after completing step 1.3. Soft tissues from the back to the front: masseter muscle (MM), soft palate (SP), hard palate (HP), gingiva (G). Hard tissue: molars and incisors. The main goal of this technique is to isolate the gingival tissue that surrounds the three molars (inside the black oval).
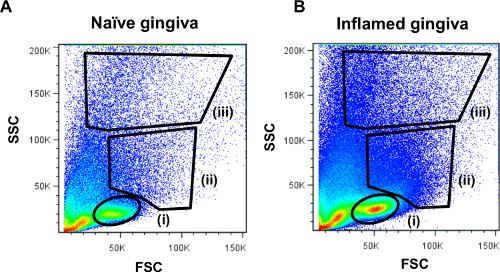
Figure 3. Representative FACS plots demonstrating cellular distribution of gingival leukocytes. Processed gingival samples were run on the LSR II flow cytometer and analyzed using FlowJo software. Side scatter (SSC) versus forward scatter (FSC) plots of the gingival cells from (A) naïve mice or (B) inflamed gingiva are presented. The location of (i) lymphocytes (ii) monocytes/dendritic cells and (iii) granulocytes populations is indicated.
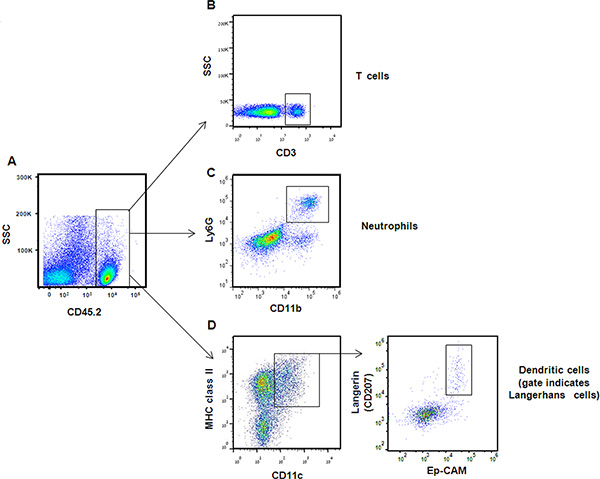
Figure 4. Representative FACS plots demonstrating analysis of immune gingival cells. (A) Immune cells were identified according to CD45 expression. Following gating on CD45-positive cells and lymphocytes population, expression of CD3 enabled targeting T-cell subset. (B) Neutrophils were identified in the gingiva according to ability of CD45+ cells to express also Ly6G and CD11b. (C) Dendritic cells were identified by gating on CD45+ cells from the monocyte/dendritic population [Figure 3, gate (ii)], and then on MHC class II and CD11c positive cells. (D) Langerhans cells were further segregated from other dendritic cells based on expression of Ep-CAM and the intracellular langerin molecule. Click here to view larger figure.
Discussion
Maxilla gingival tissues obtained from a single mouse are sufficient for analyzing sub-populations of T and B lymphocytes, as well as their capability to express extracellular and intracellular molecules as we previously described 1. Nevertheless, if rare cell populations are of interest (for example DCs), it is recommended to pool tissues from 2-3 mice. Of note, if preferable, it is possible to peel both the palatal and gingival tissues and then to excise the gingiva (a modification of step 1.8-1.9). It should be also mentioned that mandibular gingiva can be collected as well; still, its isolation is more complicated and may include other tissues that might interfere with the analysis.
Common techniques to study immune cells in the gingiva are immunohistochemistry and immunofluorescence. These techniques enable visualization of the distribution of a target molecule/cell through the gingiva, and screening of numerous fields is required in order to study a certain area. Whereas the physiological localization of certain cells cannot be determined by flow cytometry due to gingival processing, this approach provides several other advantages: (1) flow cytometry is a rapid and simple method to analyze large numbers of cells (2) it reduces analysis errors originate from tissue heterogeneity (3) it enables analysis of rare cell populations (4) it allows simultaneous analysis of various variables (5) it allows discrimination between live and dead cells (6) it permits functional analysis of the resulting cells.
The ability to perform flow cytometry analysis on gingival tissues during experimental periodontal disease enhances our ability to reveal the immunological mechanisms involved in this process. Longitudinal studying of cellular changes that occurs in the gingiva following exposure to oral pathogens should provide more relevant information rather than measuring immune responses systemically. This aspect is of particular importance when analyzing the phenotype and kinetics of innate and adaptive inflammatory cells in inflamed gingiva.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This research was supported by grants from the Israel Science Foundation (No. 1418/11) to AHH and (No. 1933/12) to AW, the German Israeli Foundation for young investigators (GIF Young) to AHH, and the Dr. I. Cabakoff Research Endowment Fund at the Hebrew University-Hadassah School of Dental Medicine to AHH and AW.
Materials
| Comments (optional) | Catalogue number | Company | Name of the reagent |
| CLS-2 | Worthington Biochemical Corp. | Collagenase Type II | |
| DN25-1G | SIGMA | DNAse I | |
| E6758-100G | SIGMA | EDTA | |
| D8537 | SIGMA | Dulbecco’s PBS | |
| Heat Inactivated | 04-121-1 | Biological Industries | Fetal Bovine Serum |
| FPE-204-500 | Jet Biofil | Vacuum-Driven Filter | |
| 352052 | BD Falcon | 5 ml Polystyrene Round-Bottom Tube | |
| 93070 | SPL Lifesciences | Cell Strainer 70 μm | |
| 153066 | NUNC | Tissue Culture Dish 35×10 mm | |
| 554714 | BD | BD Cytofix/Cytoperm | |
| Clone N418 | 117305 | Biolegend | Anti-mouse CD11c antibody |
| Clone 104 | 109819 | Biolegend | Anti-mouse CD45.2 antibody |
| Clone GK1.5 | 100413 | Biolegend | Anti-mouse CD4 antibody |
| Clone 53-6.7 | 100733 | Biolegend | Anti-mouse CD8a antibody |
| Clone 17A2 | 100214 | Biolegend | Anti-mouse CD3 antibody |
| Clone G8.8 | 118219 | Biolegend | Anti-mouse CD326 (Ep-CAM) antibody |
| Clone 929F3.01 | DDX0362D | Imgenex | Anti-mouse CD207 (Langerin) antibody |
| Clone 39-10-8 | 115010 | Biolegend | Anti-mouse I-Ad (MHC-II) antobody |
| BD Biosciences | LSR II Flow Cytometer | ||
| Tree Star | FlowJo Software v 7.6.5 |
References
- Arizon, M., et al. Langerhans cells down-regulate inflammation-driven alveolar bone loss. Proc. Natl. Acad. Sci. U.S.A. 109, 7043-7048 (2012).
- Listgarten, M. A. Pathogenesis of periodontitis. J. Clin. Periodontol. 13, 418-430 (1986).
- Kinane, D. F. Causation and pathogenesis of periodontal disease. Periodontol. 25, 8-20 (2000).
- Jotwani, R., et al. Mature dendritic cells infiltrate the T cell-rich region of oral mucosa in chronic periodontitis: in situ, in vivo, and in vitro studies. J. Immunol. 167, 4693-4700 (2001).
- Graves, D. T., et al. Interleukin-1 and tumor necrosis factor antagonists inhibit the progression of inflammatory cell infiltration toward alveolar bone in experimental periodontitis. J. Periodontol. 69, 1419-1425 (1998).
- Kabashima, H., Yoneda, M., Nagata, K., Hirofuji, T., Maeda, K. The presence of chemokine (MCP-1, MIP-1alpha, MIP-1beta, IP-10, RANTES)-positive cells and chemokine receptor (CCR5, CXCR3)-positive cells in inflamed human gingival tissues. Cytokine. 20, 70-77 (2002).
- Moughal, N. A., Adonogianaki, E., Kinane, D. F. Langerhans cell dynamics in human gingiva during experimentally induced inflammation. J. Biol. Buccale. 20, 163-167 (1992).
- Cochran, D. L. Inflammation and bone loss in periodontal disease. J. Periodontol. 79, 1569-1576 (2008).
- Taubman, M. A., Valverde, P., Han, X., Kawai, T. Immune response: the key to bone resorption in periodontal disease. J. Periodontol. 76, 2033-2041 (2005).
- Graves, D. T., Kang, J., Andriankaja, O., Wada, K., Rossa, C. Animal models to study host-bacteria interactions involved in periodontitis. Front Oral Biol. 15, 117-132 (2012).

