Genome-wide Determination of Mammalian Replication Timing by DNA Content Measurement
Summary
We describe here a relatively fast and simple approach for mapping genome-wide mammalian replication timing, from cell isolation to the basic analysis of the sequencing results. A genomic map of a representative replication program will be provided following the protocol.
Abstract
Replication of the genome occurs during S phase of the cell cycle in a highly regulated process that ensures the fidelity of DNA duplication. Each genomic region is replicated at a distinct time during S phase through the simultaneous activation of multiple origins of replication. Time of replication (ToR) correlates with many genomic and epigenetic features and is linked to mutation rates and cancer. Comprehending the full genomic view of the replication program, in health and disease is a major future goal and challenge.
This article describes in detail the "Copy Number Ratio of S/G1 for mapping genomic Time of Replication" method (herein called: CNR-ToR), a simple approach to map the genome wide ToR of mammalian cells. The method is based on the copy number differences between S phase cells and G1 phase cells. The CNR-ToR method is performed in 6 steps: 1. Preparation of cells and staining with propidium iodide (PI); 2. Sorting G1 and S phase cells using fluorescence-activated cell sorting (FACS); 3. DNA purification; 4. Sonication; 5. Library preparation and sequencing; and 6. Bioinformatic analysis. The CNR-ToR method is a fast and easy approach that results in detailed replication maps.
Introduction
Mammalian DNA replication is tightly regulated to ensure the precise replication of each chromosome exactly once during the cell cycle. Replication occurs according to a highly regulated order — multiple large genomic regions (~Mb) replicate at the beginning of S phase (early replicating domains) whereas other genomic regions replicate later at middle or late S phase (middle and late replicating domains)1. Most of the genome replicates at the same time in all tissues (constitutive ToR domains), whereas 30% – 50% of the genome, changes its ToR between tissues2, during differentiation3,4 and to a lesser extent also during cancer transformation5. Moreover, certain genomic regions replicate asynchronously6,7,8, namely there is a difference in the ToR between the two alleles.
ToR correlates with many genomic and epigenomic features including transcription levels, GC content, chromatin state, gene density, etc.1,9. ToR is also associated with mutation rates and types10,11 and therefore unsurprisingly, perturbations of the replication program are linked to cancer12,13. The causal relationship between ToR and chromatin structure is not yet understood. It is possible that open chromatin facilitates early replication. However, an alternative model suggests that the chromatin is assembled during replication and the different chromatin regulators present at the beginning and end of S phase lead to differential packaging of early and late replicating regions1,14. We have recently shown that the ToR shapes the GC content by affecting the type of mutations that occur in different genomic regions11.
Fluorescence in situ hybridization (FISH) is the main method for measuring ToR at individual loci. It is performed simply by counting the percentage of S phase cells that exhibit single FISH signals vs. the percentage of doublets for a given allele15,16. An alternative method, consists of pulse labeling the DNA with BrdU, sorting cells according to their DNA content to multiple time points along S, immunoprecipitating DNA containing BrdU, and checking the abundance of precipitated DNA with qPCR17.
Genomic ToR mapping can be achieved by two methods. The first method is a genomic version of the BrdU-IP based method described above, in which the quantification of the amount of precipitated DNA in each fraction is done simultaneously for the entire genome through hybridization to microarrays or by deep sequencing. The second method, CNR-ToR, is based on measuring the copy number of each genomic region of S phase cells and normalizing by the DNA content in G1 cells. In this method, cells are sorted by FACS into non-replicating (G1 phase) and replicating (S phase) groups (Figure 1). Cells in G1 have the same copy number in all genomic regions and thus their DNA content should be the same. On the other hand, the DNA copy number in S depends on the ToR, since early replicating regions underwent replication in most cells and therefore their DNA content is doubled, whereas late replicating regions have not replicated yet in most cells and therefore their DNA content will be similar to that of G1 cells. Hence the S to G1 ratio of DNA content is indicative of the ToR. The amount of DNA for each genomic region is measured either by hybridization to microarrays or by deep sequencing2,8. The advantages of the CNR-ToR method will be further discussed.
This paper describes the CNR-ToR method for genomic ToR mapping as described in Figure 2. The paper discusses the fine details of the entire process from collecting cells until the basic analysis of the results and the creation of genomic ToR maps. The protocol described in this paper has been successfully performed on various cell types grown in culture. Future improvements of this protocol can lead to the mapping of the ToR in vivo and in rare cell types.
Protocol
Note: ToR can be measured only on growing, unsynchronized cells. The procedure should begin with at least 1 – 2 x 106 fast growing cells, which will usually result in ~1 x 105 cells in S phase (the rate limiting step). It is recommended to conduct each experiment using two or three replicates. The entire process of CNR-ToR can be completed within one week — two days should be dedicated to all steps up to library preparation, one to two days are needed for sequencing and an additional day is necessary for the initial data analysis.
1. Collection of Cells from Culture
NOTE: The protocol is written for cells growing in culture in 10 cm plates (containing approximately 2 – 5 x 106 cells), but can be easily adjusted to other platforms.
- For cells that were grown in suspension, proceed to fixation (section 2).
- For adherent cells, aspirate and wash the plate with 3 mL PBS without Ca2+ and Mg2+.
- Discard the PBS and incubate the cells for 5 min at 37 °C with 1 mL commercial trypsin-EDTA until cells detach.
NOTE: The duration of the trypsin treatment should be adjusted to each cell type. - Add 3 mL culture media to neutralize the trypsin and collect cells in a 15 mL conical tube or a 5 mL polystyrene tube. Keep on ice.
2. Fixation
NOTE: For this part, all steps should be done at 4 °C.
- Centrifuge the cells at 300 x g for 5 min at 4 °C.
- Aspirate and wash cells twice with 1 mL cold PBS.
- Resuspend cells in 250 µL (total) cold PBS.
- While gently vortexing the tube, slowly add dropwise 800 µL of -20 °C 100% ethanol. This leads to a final ethanol concentration of 70 – 80%.
NOTE: High purity ethanol is recommended at this step. - Incubate cells on ice for 30 min.
NOTE: At this stage cells can be kept for a few days at 4 °C or for a few months at -20 °C.
3. PI Staining
- Centrifuge the cells at 500 x g for 10 min at 4 °C.
- Aspirate the supernatant carefully and wash cells twice with 1 mL cold PBS.
- Aspirate and resuspend each sample with the following mixture: 1 mL PBS, 5 µL 10 mg/mL RNaseA, 50 µL 1 mg/mL propidium iodide (PI; mix bottle before use). Adjust concentration up to ~2 x 106 cells/mL).
NOTE: Keep out of light — PI is light sensitive. - Filter through 35 µm mesh to a 5 mL polystyrene tube, and close with Parafilm.
- Incubate at RT in the dark, for 15 – 30 min.
NOTE: Cells are now ready for FACS analysis. If necessary, the stained cells can be stored for at least 24 h at 4 °C in the dark.
4. Sort
- Sort cells using a FACS machine. Use the 561 nm laser to differentiate cells based on their PI intensity. Other lasers near the 535 nm excitation maximum like 488 nm or 532 can be used depending on the FACS machine configuration.
- For optimal results, use the smallest nozzle recommended for the specific cell size (for most cells 85 µm). Prioritize purity modes over yield. Use a steady slow flow, usually up to 300-500 events/s, with a sheath pressure of 45 psi.
- Using gating, discriminate dead cells and subcellular debris (low FCS and high SSC) by Plotting FCS vs SSC. From the viable cells discriminate doublets by plotting SSC-Width (W) vs SSC-Height (H) followed by a FSC-W vs FSC-H plot and by PI- W vs PI-H (doublets will have the same H-value but bigger W-value). For the viable single cells draw a histogram of the PI-Area (A) intensity which represents the DNA content of the cells.
- Sort cells into G1 and S phases, as shown in Figure 1. Gating for S should be wide and intrude into the G1 and G2 phases, while G1 gating should be narrow and as far from S as possible.
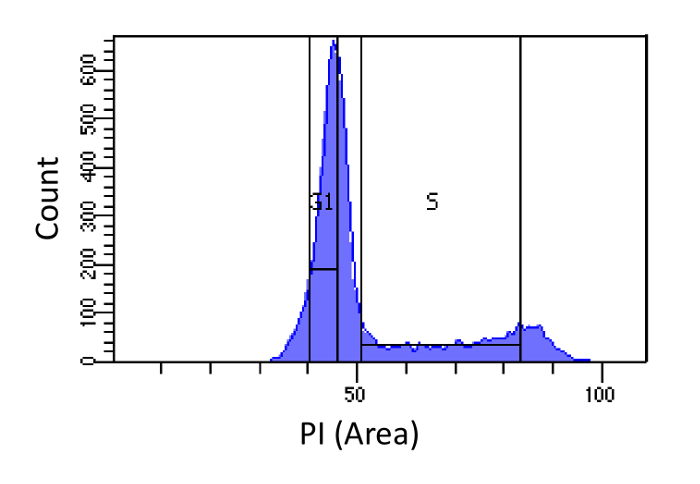
Figure 1. Cell cycle phase determination based on PI intensity. Histogram showing the distribution of the cellular DNA content (measured by PI-Area) of mouse embryonic fibroblast (MEF) population. The DNA content is used to sort the population into two sub-populations i) G1 cells (2N DNA content) and ii) S phase cells (2N – 4N DNA content), using the marked regions. Please click here to view a larger version of this figure.
NOTE: The purpose of the collection of G1 cells is to account for biases in the sequencing efficiency between different genomic regions. An alternative approach is to use G1 arrested cells from the same cell type. This approach gives cleaner results (since it minimizes S phase contamination) but it may introduce biases stemming from genetic differences between the arrested cells and the measured cells.
- Sort in cold conditions and collect sorted cells into 1.5/2 mL tubes. Keep tubes on ice following the sort.
NOTE: In order to improve DNA recovery, it is better to use low binding tubes or to use tubes coated with 4 – 5% BSA for 1 – 3 h at 4 °C 18.
5. DNA Purification
- For each sample (G1 and S) purify DNA using a DNA purification kit.
NOTE: When using the commercial kit, elute with 400 µL elution buffer into a 2 mL tube for high yield, as recommended by the manufacturer. - Check DNA concentration using fluorometer.
NOTE: From 100,000 mammalian cells collected by the FACS, one should get ~ 1 µg of DNA. At this stage DNA can be kept for a few days at 4 °C or at -20 °C for long storage.
6. Sonication
- Transfer DNA into a 1.7 mL tube that is compatible with the magnetic stand used.
- Concentrate DNA using 2x SPRI beads according to the manufacturer's instructions, using a magnetic stand, and elute in 50 µL elution buffer.
- Shear DNA with a focused ultrasonicator to an average target peak size of 250 bp. Use the following settings for a 50 µL DNA sample: 50 W, 20% Duty factor, 200 Cycles per burst, 20 °C, 120 s.
- Verify the size of the sheared DNA by electrophoresis. The recommended size distribution is 200 – 700 bp with a peak at ~ 250 bp.
NOTE: At this stage DNA can be kept for a few days at 4 °C or at -20 °C for long storage.
7. Library Preparation, and Sequencing
NOTE: Many library preparation kits and different sequencing platforms should work similarly to the ones used by us and mentioned in the materials section. Actually in the past, ToR maps were generated using a very similar method with microarray platforms2.
- Prepare libraries using any commercial library preparation kit.
- At the end of library preparation, select size using magnetic beads for 300 – 800 bp.
- After preparing libraries, measure DNA concentration using fluorometer.
- Measure DNA size using electrophoresis.
- Perform sequencing on any platform.
NOTE: Sequencing at least 10 M reads per sample is recommended. This depth is equivalent to a read approximately every 300 bp (for ~3 Gb size genomes), and is sufficient for ToR measurements at a resolution of 50 – 100 kb. Increasing the depth will result in a decrease in the size of the windows and thus will enable increasing resolution with higher certainty. Paired end sequencing is not necessary in this protocol since only coverage information is collected. It, however, may help in resolving the location of reads that contain repetitive sequences.
8. Analysis
NOTE: Data analysis is based on the method used by A. Koren et al.19.
- Map sequencing data to the corresponding genome using bowtie2 or any reliable short read aligner. Define varying-size, equal-coverage chromosomal windows as segments covered by 200 reads in the G1 fraction and count S phase reads in the same windows.
- Calculate the S/G1 ratio for each window. This should generate a map with large fluctuations in the S/G1 ratio along the genome (Figure 3). A good control for the reliability of the ToR measurements is to compare this map to the G1/G1 ratio (from two separate measurements of G1) which should be much flatter.
- Normalize data to 0 mean and 1 SD by subtracting from each value the average value of all windows (excluding the X chromosome) and dividing the results by the standard deviation of the S/G1 of all windows. This is done in order to convert to z scores and enable comparison between different experiments.
- Remove all gap regions listed by the UCSC genome browser as well as each remaining inter gap fragment containing fewer than 15 data windows.
- Smooth the remaining fragments with a cubic smoothing spline via the Matlab function csaps with a parameter of 10−16 and interpolate at set points every 100 kb.
NOTE: Parameters of smoothing and interpolation should be adjusted based on the depth of the data. Other suitable smoothing methods and functions exist and can be used. - After visually confirming the reliability of each replicate, merge all the reads and calculate a deeper resolution profile by performing the same process described above on this data.
Representative Results
A typical ToR map is shown in Figure 3 for mouse embryonic fibroblasts (MEFs). This figure demonstrates the analysis process since it shows both the dots, which are the normalized S/G1 ratio for individual windows (step 8.3), as well as the line which results from the cubic smoothing and interpolation (step 8.5).
Such maps capture the organization of the replication program, which is a patchwork of two types of ToR domains: i) large regions (in the order of a megabase) which replicate simultaneously (CTRs = constant ToR regions) at early, middle or late S; and ii) temporal transition regions (TTRs) in which the ToR changes gradually. CTRs are connected to each other by TTRs and together these two types of replication organization cover the entire genome.
Despite the relatively low sequencing coverage (one read every 300 bp) used for ToR mapping, the resulting maps are quite robust. Figure 4 shows the reproducibility of ToR maps between triplicates of MEF cells compared to triplicates of mouse pre-B cells in the same region. Comparison between such maps allows the identification of regions with differential ToR which are defined as regions in which the differences in the ToR maps between tissues are significantly larger than the differences between replicates.
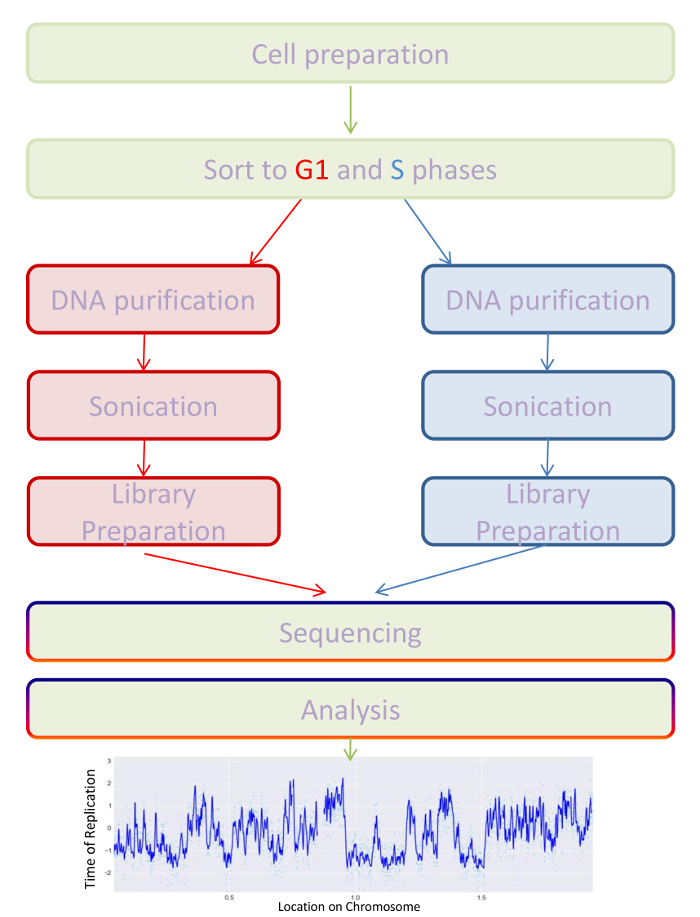
Figure 2: Protocol scheme. Schematic diagram describing the procedure of the CNR-ToR method. Please click here to view a larger version of this figure.
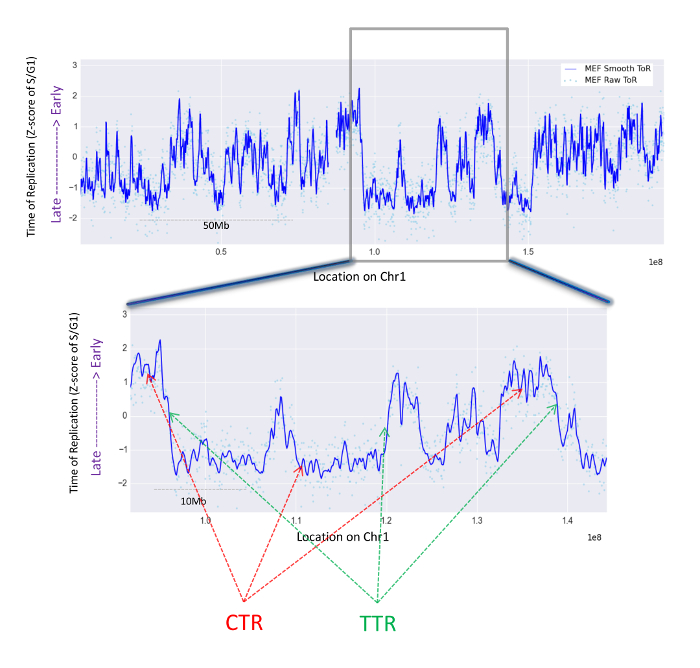
Figure 3: Representative genomic ToR maps. Shown is the ToR of the entire Chromosome 1 of MEF cells and a closer look at a ~50 Mb region. Shown are the Z scores of the S/G1 values in varying size windows (dots) and the smoothed data (solid line). High S/G1 values correspond to early replication whereas low values correspond to late replication. Arrows point to examples of constant ToR regions (CTRs; red) and temporal transition regions (TTRs; green). Please click here to view a larger version of this figure.
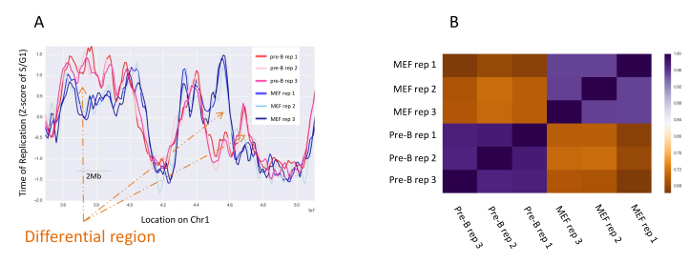
Figure 4: Reproducibility of the repeats. (A) Smoothed ToR of MEF triplicates (blue) compared to pre-B triplicates (red) in an 18 Mb region on mouse chromosome 1. Orange arrows point to examples of differential regions between the cell lines. (B) Heat map of the spearman correlations between the 6 samples. Please click here to view a larger version of this figure.
Discussion
CNR-ToR can be performed in principle on any eukaryotic proliferating cell population that can be divided by FACS to S and G1 phases (reviewed by Rhind N. and Gilbert DM20). The method described here has been adjusted to mammalian cells with a genome size of ~3 Gb such as human and mouse. Small changes in the CNR-ToR protocol (in cell preparation and sequencing depth) are needed, in order to adjust it to other eukaryotes. Attention must be paid to the collection of sufficient amounts of S phase cells since it is the rate-limiting step. Thus, a preliminary FACS for cell cycle should be performed to confirm that the experiment can be done and to validate the amount of cells in S phase. For cells which exhibit irregular cell cycles, it is recommended to prestain the cells with BrdU to detect dividing cells, and follow the Anti-BrdU manufacturer protocol until the FACS step. Usually cells are stained with 10-20 µM BrdU for 30 min.
It is recommended to perform each experiment in biological triplicates to detect inconsistencies that may stem from differences in cell growth and in the sorting process. This is especially important for the identification of small changes in the ToR maps between cell types and conditions. For fast growing cells, a single 10 cm tissue culture plate/flask (containing approximately 2-5 x 106 cells) is recommended for each replicate in order to get >1 x 105 cells in S phase. This amount should be increased when working with slow growing cells, since the percentage of cells in S phase is lower and thus one has to sort more cells in order to get a sufficient amount of cells in S.
When sorting cells, it is important to achieve the best possible separation of the cells according to their DNA content and thus a low flow rate is recommended. In order to maximize the temporal resolution, it is important to collect cells from the entire S phase. This is achieved by gating S as wide as possible (Figure 1). On the other hand, G1 gating should be narrow (starting from the G1 peak and to the left; Figure 1), in order to avoid both sub-G1 and early S-phase contaminations.
Sonication can be performed by various methods, but it is recommended to use a focused ultrasonicator which enables a robust and easy sonication without the need for calibration of each experiment. Nevertheless, when first establishing the protocol in the laboratory, it is recommended to calibrate the parameters for obtaining a size distribution of 200-700 bp.
As explained in the introduction, there are two main methods for genome-wide ToR mapping — BrdU-IP and CNR-ToR. Both methods give similar ToR maps (data not shown) despite the differences between them. The CNR-ToR method is limited in its enrichment range since the maximum difference between early and late regions is twofold, whereas with the BrdU-IP much higher enrichment is achieved, since early replicating regions will contain BrdU almost only in the early fraction. On the other hand, the BrdU-IP method resolution is limited by the number of S phase fractions collected. In its most common application, only two fractions (early versus late S) are compared, which results in a compromise of the fine temporal resolution. The CNR-ToR method, however, gives a continuous signal along the entire S phase. Moreover, the BrdU-IP method is based on immuno-precipitation which usually gives a much higher background than the CNR-ToR method. Another advantage of the CNR-ToR method is that it can be used retrospectively to extract ToR information from the sequencing data of unsynchronized cultures as recently described21. Finally, the CNR-ToR method is preferable due to its simplicity and due to the fact that it can be down-scaled since it is not based on immuno-precipitation.
The role of ToR in the complex network of genomic and epigenomic features remains to be deciphered. The relative ease of the CNR-ToR method allows for the expansion of the existing ToR data to include many of the natural conditions that cells experience and should be explored thoroughly and genome wide. This includes various replication stress situations, endoreduplication as well as various cancer transformations. The use of SNP data and higher sequencing depth will also allow measuring the ToR of each allele separately. Moreover, the further decrease in sequencing cost will enable increasing of the resolution of ToR maps, which may further aid in allow the identification of subtle changes between conditions. Other future applications may be achieved by improving the current methods and applying ToR measurement on small numbers of cells which will enable measuring ToR in vivo.
Divulgations
The authors have nothing to disclose.
Acknowledgements
We thank Oriya Vardi for assistance in generating figures. Work in the IS group was supported by the Israel Science Foundation (grant No. 567/10) and the European Research Council Starting Grant (#281306).
Materials
| PBS | BI (Biological Industries) | 02-023-1A |
| Trypsin-EDTA | BI (Biological Industries) | 03-052-1B |
| 15ml conical tube | Corning | 430790 |
| 5ml Polystyrene round Bottom tube with cell strainer cap | BD-Falcon | 352235 |
| Ethanol | Gadot | 64-17-5 |
| RNAse-A 10mg/ml | Sigma | R4875 |
| Propidiom iodide 1mg/ml | Sigma | P4170 |
| parafilm | Parafilm | PM-996 |
| 1.5ml DNA LoBind Eppendorf tubes | Eppendorf | 22431021 |
| BSA | Sigma | A7906 |
| 1.7ml MaxyClear tube | Axygen | MCT-175-C |
| magnetic beads – Agencourt AMPure XP | Beckman Coulter | A63881 |
| Ultrasonicator | Covaris | M-series -530092 |
| 50 µl microTUBE AFA Fiber Screw-Cap 6x16mm | Covaris | 520096 |
| Qubit fluorometer | Invitrogen | |
| Qubit dsDNA High Sensitivity (HS) Assay Kit | Invitrogen | Q32854 |
| Electrophoresis.2200 Tape station system | Agilent | D1000 ScreenTape |
| Seqeuncing – Illumina NextSeq system | Illumina | SY-415-1001 |
| Dneasy kit for DNA purification | Qiagen | 69504 |
| PureProteom Magnetic Stand | Millipore | LSKMAGS08 |
| Anti-BrdU/FITC | DAKO | F7210 |
| FACS sorter | BD | FACSARIA III |
| FACS software | BD | FACSDiva v 8.0.1 |
References
- Farkash-Amar, S., Simon, I. Genome-wide analysis of the replication program in mammals. Chromosome Res. 18 (1), 115-125 (2010).
- Yaffe, E., et al. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS Genet. 6 (7), e1001011 (2010).
- Hiratani, I., et al. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 6 (10), (2008).
- Rivera-Mulia, J. C., et al. Dynamic changes in replication timing and gene expression during lineage specification of human pluripotent stem cells. Genome Res. 25 (8), 1091-1103 (2015).
- Ryba, T., et al. Abnormal developmental control of replication-timing domains in pediatric acute lymphoblastic leukemia. Genome Res. 22 (10), 1833-1844 (2012).
- Farkash-Amar, S., et al. Global organization of replication time zones of the mouse genome. Genome Res. 18 (10), 1562-1570 (2008).
- Koren, A., McCarroll, S. A. Random replication of the inactive X chromosome. Genome Res. 24 (1), 64-69 (2014).
- Mukhopadhyay, R., et al. Allele-specific genome-wide profiling in human primary erythroblasts reveal replication program organization. PLoS Genet. 10 (5), e1004319 (2014).
- McNairn, A. J., Gilbert, D. M. Epigenomic replication: linking epigenetics to DNA replication. Bioessays. 25 (7), 647-656 (2003).
- Sima, J., Gilbert, D. M. Complex correlations: replication timing and mutational landscapes during cancer and genome evolution. Curr Opin Genet Dev. 25, 93-100 (2014).
- Kenigsberg, E., et al. The mutation spectrum in genomic late replication domains shapes mammalian GC content. Nucleic Acids Res. 44 (9), 4222-4232 (2016).
- Woo, Y. H., Li, W. H. DNA replication timing and selection shape the landscape of nucleotide variation in cancer genomes. Nat Commun. 3, 1004 (2012).
- Liu, L., De, S., Michor, F. DNA replication timing and higher-order nuclear organization determine single-nucleotide substitution patterns in cancer genomes. Nat Commun. 4, 1502 (2013).
- Goren, A., Cedar, H. Replicating by the clock. Nat Rev Mol Cell Biol. 4 (1), 25-32 (2003).
- Selig, S., Okumura, K., Ward, D. C., Cedar, H. Delineation of DNA replication time zones by fluorescence in situ hybridization. EMBO J. 11 (3), 1217-1225 (1992).
- Smith, L., Thayer, M. Chromosome replicating timing combined with fluorescent in situ hybridization. J Vis Exp. (70), e4400 (2012).
- Simon, I., et al. Asynchronous replication of imprinted genes is established in the gametes and maintained during development. Nature. 401 (6756), 929-932 (1999).
- Phi-Wilson, J. T., Recktenwald, D. J. Coating agents for cell recovery. Google Patents. , (1993).
- Koren, A., et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. Am J Hum Genet. 91 (6), 1033-1040 (2012).
- Rhind, N., Gilbert, D. M. DNA replication timing. Cold Spring Harb Perspect Biol. 5 (8), a010132 (2013).
- Koren, A., et al. Genetic variation in human DNA replication timing. Cell. 159 (5), 1015-1026 (2014).

