Dissection of Human Retina and RPE-Choroid for Proteomic Analysis
Summary
The human retina is composed of functionally and molecularly distinct regions, including the fovea, macula, and peripheral retina. Here, we describe a method using punch biopsies and manual removal of tissue layers from a human eye to dissect and collect these distinct retinal regions for downstream proteomic analysis.
Abstract
The human retina is composed of the sensory neuroretina and the underlying retinal pigmented epithelium (RPE), which is firmly complexed to the vascular choroid layer. Different regions of the retina are anatomically and molecularly distinct, facilitating unique functions and demonstrating differential susceptibility to disease. Proteomic analysis of each of these regions and layers can provide vital insights into the molecular process of many diseases, including Age-Related Macular Degeneration (AMD), diabetes mellitus, and glaucoma. However, separation of retinal regions and layers is essential before quantitative proteomic analysis can be accomplished. Here, we describe a method for dissection and collection of the foveal, macular, and peripheral retinal regions and underlying RPE-choroid complex, involving regional punch biopsies and manual removal of tissue layers from a human eye.One-dimensional SDS-PAGE as well as downstream proteomic analysis, such as liquid chromatography-tandem mass spectrometry (LC-MS/MS), can be used to identify proteins in each dissected retinal layer, revealing molecular biomarkers for retinal disease.
Introduction
The retina, RPE, and choroid are complex tissues that demonstrate important regional differences in protein expression, physiological function, and pathological susceptibilities1,2. For example, diseases such as Age-Related Macular Degeneration (AMD), retinitis pigmentosa, and central serous retinopathy each demonstrate characteristic localization within the fovea, macula, or retina periphery1,3,4,6. Here, we present a method demonstrating how distinct retinal regions can be independently sampled. The overall goal of this method is to provide a reliable guide for collection of tissue samples from the foveal, macular, and peripheral regions of the human retina and RPE-choroid for proteomic analysis. The rationale for the development and use of this technique is that through proteomic analysis of these specific retinal regions, important molecular insights may be gained into the physiological and pathophysiological functions of these regions.
This approach promises to reveal the proteomic basis for relative regional disease susceptibilities, and to facilitate the identification of new specific therapeutic targets. Indeed, proteomic investigations of the vitreous and its interactions with the retina have provided key insights into the molecular composition and function of healthy and diseased tissue5,7,8,9,10,11,12,13. However, clear comparative proteomic analyses of distinct retinal regions are lacking. The technique will help to support these much-needed studies, providing advantages over other methods by demonstrating a reliable and reproducible tissue collection approach. More so, the approach is very accessible, taking advantage of standard-sized and readily available tissue punch biopsy tools. Our technique emphasizes the appropriate collection and storage of tissues for proteomic processing, making important considerations for protein stability and degradation. Thus, this method is most appropriate for investigators considering downstream molecular analysis of proteomic factors.
Protocol
This study was approved by the University of Iowa's Institutional Review Board and adheres to the tenets set forth in the Declaration of Helsinki.
1. Foveal and Macular Biopsy Punch
- Open and butterfly the human eye, such that it consists of 4 separate flaps of tissue, as described in a previous publication.5
- Beginning with a butterflied human eye placed in a Petri dish, center a 4-mm punch biopsy tool over the fovea, press down, and roll gently until an incision is made around the fovea.
- Next, generate an incision around the macula by centering and pressing down an 8-mm skin punch biopsy tool within the arcades of the macula, applying gentle pressure and rolling. This will produce a second, outer ring of tissue surrounding the first.
2. Peripheral Retinal Biopsy Punch
- Use the 4-mm skin punch biopsy tool to make a series of punches in the peripheral retina, just outside the arcade. Here, make two punches for each quadrant flap.
NOTE: After all the punches are made, the eye will have two concentric punches in the center, representing the fovea and macula, and two punches at the base of each flap-totaling 8 punches in the peripheral retina.
3. Fovea and Macula Biopsy Collection
- As the tissue is now ready for collection, collect all tissue biopsies in separate microfuge tubes and freeze using liquid nitrogen for downstream processing. Store all samples at -80 °C until utilization.
- Use a curved 0.12 Colibri forceps to grab the edges of the translucent tissue of the retinal fovea. To collect, elevate and separate the foveal tissue from the underlying RPE-choroid.
- Similarly, use the 0.12 Colibri forceps to grasp the outer ring of the translucent macula tissue. If the tissue is still attached to underlying or adjacent tissue, use a pair of Westcott scissors to carefully trim the edge, capturing just the retinal component and dissecting away any optic nerve tissue included in the punch.
4. Peripheral Retina Collection
- Use the curved 0.12 Colibri forceps to gently separate the peripheral retinal tissue discs from the underlying RPE-choroid and place within individual microfuge tubes.
- In the case of residual vitreous gel, use the forceps to lift and separate the gel away as much as possible before collection of the retinal tissue. Once the retina has been removed, the pigmented RPE-choroid remains below.
5. Foveal and Macula RPE-Choroid Collection
- Use the 0.12 Colibri forceps to grasp the edges of the dark RPE-choroid tissue that lies beneath the area of the removed fovea. Carefully separate this tissue from the sclera and collect.
- Using the same forceps, grasp the edges of the outer ring of dark RPE-choroid tissue underlying the removed macular area. Carefully separate this tissue from the sclera by grasping the edges at different points around the ring and lightly pulling. Eventually, the ring of RPE-choroid tissue will be dislodged and may be collected.
NOTE: Secondary forceps in the opposite hand can be helpful in manipulating the RPE-choroid tissue during removal. Like the retinal tissue, Wescott scissors can be used to assist in removing any tissue that was not fully incised using the punch tool.
6. Peripheral Retina RPE-Choroid Collection
- Use the 0.12 Colibri forceps to peel away the RPE-choroid in the 8 peripheral punch areas.
- As previously, place the RPE-choroid tissue in microfuge tubes and freeze using liquid nitrogen for downstream processing. Keep all samples at -80 °C until utilization.
7. Scissor Dissection
NOTE: If the punch biopsy blade is dull or the punch biopsy tool is not pushed hard enough, there may not be a clean-cut surrounding the tissue.
- In these cases, pull the tissue away as much as possible, and then use Westcott scissors to trim and separate.
Representative Results
Retinal and RPE-choroid tissue can be processed in various ways to suit an individual investigation. After collection, the researcher will possess samples of retinal and RPE-choroid tissue from the foveal region, outer macula, and peripheral retina (Figure 1). Specifically, the foveal region punch will include the fovea, the parafovea, and a small amount of the adjacent perifovea. The macular punch includes the remainder of the perifoveal region as well as a small amount of the adjacent near-peripheral region. Finally, the peripheral punches sample the mid-peripheral and far-peripheral regions. In a representative experiment, tissue samples were trypsin-digested and analyzed using one-dimensional SDS-PAGE to visualize protein content (Figure 2A). The results of this analysis suggest distinct proteins among the different regions of the retina. Analysis by liquid chromatography-tandem mass spectrometry (LC-MS/MS)6 properly identified peptides from rhodopsin, a highly abundant and unique retina protein. A representative rhodopsin spectrum obtained from the macular region is shown in Figure 2B. Further analysis of the protein content will provide insights into the molecular functions of the anatomically distinct regions of the retina. In the case that the dissection is not performed carefully, and the retina is not properly separated from the RPE-choroid, differences in protein content between these two tissues will not be discernable.
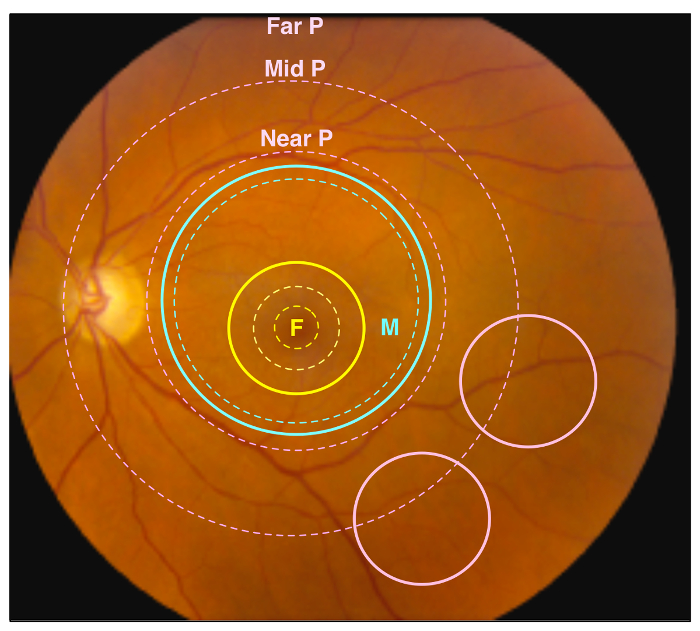
Figure 1. Retinal Regions. A representative image of a healthy human retina. Different retinal regions are highlighted by dotted circles, and regions sampled by punch biopsies are indicated by solid circles. The yellow dotted circles represent the foveal region, including the fovea, parafovea, and perifovea (traveling outward from center), while the yellow solid circle represents the 4 mm foveal punch. The blue dotted circle represents the anatomic macular region, while the blue solid circle represents the 8 mm macular punch. Finally, the pink dotted circles represent the near retinal peripheral region (Near P), mid retinal peripheral region (Mid P), and far peripheral region (Far P). The pink solid circles represent the 4 mm peripheral retina punches, which contain the Mid P and Far P regions. Please click here to view a larger version of this figure.
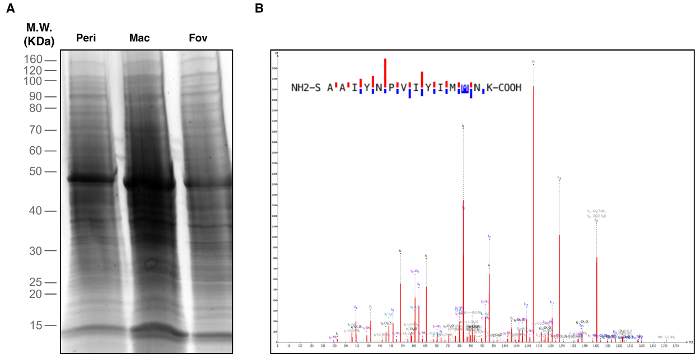
Figure 2. Identification of Retinal Proteins. Peripheral retina, macula, and foveal regions were biopsied and used for proteomic analysis. (A) One-Dimensional SDS-PAGE and silver staining visualized proteins in each retinal region. (B) Retina tissue samples were subject to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The representative spectrum shown is of the rhodopsin protein identified in the macular region. This spectrum represents one of five unique rhodopsin peptides identified in the macula. Please click here to view a larger version of this figure.
Discussion
After tissue collection, sample handling and treatment are crucial considerations14. Preservation in liquid nitrogen is preferred over chemical fixation, as the latter may result in damage to protein structure, which could skew downstream analysis. Additionally, liquid nitrogen preservation is preferred to methods which do not involve freezing of samples. Notably, Ferrer et al. showed significant differences in protein levels between brain samples preserved at 4 °C or room temperature, compared to those stored at 0 °C15. Further, it is important to be careful when processing samples, as specific chemical and physical treatment before freezing has the potential to alter protein levels and post-translational modifications16,17. This point emphasizes the importance of considering the intended downstream analysis of samples. Separately, post-mortem timing of tissue harvesting is another factor that can affect levels and stability of specific proteins. Degradation can occur to the sample proteome depending on time post-mortem and storage temperature18,19,20. Moreover, controlling for time post-mortem degradation, such as by use of internal controls between disease tissue and healthy tissue, is crucial. If the timing factor is ignored, degradation could skew results. As a general rule, the sooner the tissue is analyzed post-mortem, the less risk for degradation to alter study results.
Several procedural modifications can be made to this protocol to better suit a specific research question. One modification is that different size skin punch biopsy tools may be used to collect different amounts of tissue samples. Less or more tissue may be required for downstream proteomic processing depending on quantity or stability of specific proteins of interest. Moreover, the size of the tissue punch biopsy can determine how grossly the specific regions of the retina or RPE-choroid will be sampled. For example, if specific, fine regions of pathologic tissue are required, smaller skin punch biopsy tools may be needed. Furthermore, if a section is so fine that it drops below the capabilities of punch biopsy tools, microdissection may be required21. Additionally, the use of visual aids, such as a dissecting scope or magnifying glass, may aid the tissue collection process-particularly in cases of smaller globe sizes, such as with infantile ocular tissue.
Finally, it is important to note that while this technique groups the RPE and choroid tissues as a single complex, these tissues remain molecularly and functionally distinct. Indeed, the proteomes differ markedly between these two tissues. However, despite these differences, the RPE and choroid remain anatomically, functionally, and clinically linked2,6,7. Thus any downstream proteomic analysis of the RPE-choroid complex should bear in mind this key distinction.
This manuscript demonstrates a straightforward, reliable, low-overhead approach to sample collection for proteomic analysis of fovea, macula, and peripheral retina and RPE-choroid tissue in human eyes. At the discretion of the investigator, modifications can be made to either the collection process or tissue handling, pre-collection and post-collection, depending on the intended downstream proteomic analysis.
Divulgations
The authors have nothing to disclose.
Acknowledgements
VBM is supported by NIH grants [K08EY020530, R01EY024665, R01EY025225, R01EY024698 and R21AG050437], Doris Duke Charitable Foundation Grant #: 2013103, and Research to Prevent Blindness (RPB), New York, NY. MT and GV are supported by NIH grant T32GM007337.
Materials
| 4-mm skin punch biopsy tool | Miltex | REF 33-34 | |
| 8-mm skin punch biopsy tool | Miltex | REF 33-37 | |
| 0.12 Colibri Forceps | Stephens Instruments | S5-1145 | |
| Wescott Scissors | Sklar Surgical Instruments | 64-3146 | |
| Microfuge tubes | Eppendorf | #022364111 | 1.5 mL |
| Liquid Nitrogen | Praxair, Inc. | 7727-37-9 [R] |
References
- Chirco, K. R., Sohn, E. H., Stone, E. M., Tucker, B. A., Mullins, R. F. Structural and molecular changes in the aging choroid: implications for age-related macular degeneration. Eye (Lond). , (2016).
- Zhang, P., et al. Defining the proteome of human iris, ciliary body, retinal pigment epithelium, and choroid. Proteomics. 16 (7), 1146-1153 (2016).
- Funke, S., et al. Glaucoma related Proteomic Alterations in Human Retina Samples. Sci Rep. 6, 29759 (2016).
- Decanini, A., et al. Human retinal pigment epithelium proteome changes in early diabetes. Diabetologia. 51 (6), 1051-1061 (2008).
- Skeie, J. M., Mahajan, V. B. Dissection of human vitreous body elements for proteomic analysis. J Vis Exp. (47), (2011).
- Skeie, J. M., Mahajan, V. B. Proteomic landscape of the human choroid-retinal pigment epithelial complex. JAMA Ophthalmol. 132 (11), 1271-1281 (2014).
- Skeie, J. M., Tsang, S. H., Mahajan, V. B. Evisceration of mouse vitreous and retina for proteomic analyses. J Vis Exp. (50), (2011).
- Skeie, J. M., et al. Proteomic analysis of vitreous biopsy techniques. Retina. 32 (10), 2141-2149 (2012).
- Skeie, J. M., Mahajan, V. B. Proteomic interactions in the mouse vitreous-retina complex. PLoS One. 8 (11), e82140 (2013).
- Mahajan, V. B., Skeie, J. M. Translational vitreous proteomics. Proteomics Clin Appl. 8 (3-4), 204-208 (2014).
- Skeie, J. M., Roybal, C. N., Mahajan, V. B. Proteomic insight into the molecular function of the vitreous. PLoS One. 10 (5), e0127567 (2015).
- Velez, G., et al. Precision Medicine: Personalized Proteomics for the Diagnosis and Treatment of Idiopathic Inflammatory Disease. JAMA Ophthalmol. 134 (4), 444-448 (2016).
- Velez, G., et al. Proteomic analysis of elevated intraocular pressure with retinal detachment. Am J Ophthalmol Case Rep. 5, 107-110 (2017).
- Skeie, J. M., et al. A biorepository for ophthalmic surgical specimens. Proteomics Clin Appl. 8 (3-4), 209-217 (2014).
- Ferrer, I., et al. Brain protein preservation largely depends on the postmortem storage temperature: implications for study of proteins in human neurologic diseases and management of brain banks: a BrainNet Europe Study. J Neuropathol Exp Neurol. 66 (1), 35-46 (2007).
- Ahmed, M. M., Gardiner, K. J. Preserving protein profiles in tissue samples: differing outcomes with and without heat stabilization. J Neurosci Methods. 196 (1), 99-106 (2011).
- Kanshin, E., Tyers, M., Thibault, P. Sample Collection Method Bias Effects in Quantitative Phosphoproteomics. J Proteome Res. 14 (7), 2998-3004 (2015).
- Crecelius, A., et al. Assessing quantitative post-mortem changes in the gray matter of the human frontal cortex proteome by 2-D DIGE. Proteomics. 8 (6), 1276-1291 (2008).
- Oka, T., Tagawa, K., Ito, H., Okazawa, H. Dynamic changes of the phosphoproteome in postmortem mouse brains. PLoS One. 6 (6), e21405 (2011).
- Nagy, C., et al. Effects of postmortem interval on biomolecule integrity in the brain. J Neuropathol Exp Neurol. 74 (5), 459-469 (2015).
- Mukherjee, S., et al. Proteomic analysis of frozen tissue samples using laser capture microdissection. Methods Mol Biol. 1002, 71-83 (2013).

