SUMO-Binding Entities (SUBEs) as Tools for the Enrichment, Isolation, Identification, and Characterization of the SUMO Proteome in Liver Cancer
Summary
Here, we present a protocol to enrich, isolate, identify, and characterize proteins modified by SUMO in vivo both from human hepatoma cells and liver tumors obtained from mouse models of hepatocellular carcinoma by using SUMO-binding entities (SUBEs).
Abstract
Post-translational modification is a key mechanism regulating protein homeostasis and function in eukaryotic cells. Among all ubiquitin-like proteins in liver cancer, the modification by SUMO (Small Ubiquitin MOdifier) has been given the most attention. Isolation of endogenous SUMOylated proteins in vivo is challenging due to the presence of active SUMO-specific proteases. Initial studies of SUMOylation in vivo were based on the molecular detection of specific SUMOylated proteins (e.g., by western blot). However, in many cases, antibodies, generally made with non-modified recombinant protein, did not immunoprecipitate SUMOylated forms of the protein of interest. Nickel chromatography has been the other approach to study SUMOylation by capturing histidine-tagged versions of SUMO molecules. This approach is mainly used in cells stably expressing or transiently transfected with His-SUMO molecules. To overcome these limitations, SUMO-binding entities (SUBEs) were developed to isolate endogenous SUMOylated proteins. Herein, we describe all the steps required for the enrichment, isolation, and identification of SUMOylated substrates from human hepatoma cells and hepatic tissues from a liver cancer mouse model by using SUBEs. Firstly, we describe methods involved in the preparation and lysis of the human hepatoma cells and liver tumor tissue samples. Then, a thorough explanation of the preparation of SUBEs and controls is detailed along with the protocol for the protein pull-down assays. Finally, some examples are provided regarding the options available for the identification and characterization of the SUMOylated proteome, namely the use of western-blot analysis for the detection of a specific SUMOylated substrate from liver tumors or the use of proteomics by mass spectrometry for high-throughput characterization of the SUMOylated proteome and interactome in hepatoma cells.
Introduction
Liver cancer is the sixth most common cancer worldwide and the second cause of cancer-associated deaths1. Hepatocellular Carcinoma (HCC) is the most prevailing form of primary liver cancer. Historically, common risk factors for the development of HCC included chronic hepatitis B or C infection and abusive alcohol consumption. In the last decades, the metabolic syndrome, Type 2 diabetes non-alcoholic fatty liver disease (NAFLD) has emerged as risk factors for the development of HCC2. HCC is very heterogeneous, both phenotypically and genetically, wherein a complex network of signaling pathways are disrupted. In the last years, even though there has been an increase in our knowledge about the molecular pathways implicated in the pathogenesis of HCC, there are still no effective therapeutic approaches for HCC management. Many pathways are activated in HCC and inhibiting one generally drives the compensation by other pathways3. This has been one of the main difficulties when treating HCC. Thus, a more global approach may provide a potential therapeutic approach for the clinical management of liver cancer e.g., targeting post-translational modifications (PTMs), as multiple signaling pathways can be simultaneously regulated by PTMs of proteins.
Post-translational modifications are considered as key mechanisms regulating protein homeostasis and functions4. Structural and functional changes are introduced by PTMs, thereby, increasing proteome diversity. The most common PTMs include phosphorylation, methylation, acetylation, glycosylation, ubiquitination, and conjugation of ubiquitin-like proteins (UbLs). Among all UbLs, protein modification by SUMO (Small Ubiquitin MOdifier) has attracted attention in association with its critical role in a variety of cellular processes, including transcription, cellular localization, DNA repair, and cell cycle progression5. Recently, SUMOylation was shown to be altered in liver cancer6,7,8,9, and changes in the SUMOylation of specific proteins has been described to play a role in the progression of cancer-related diseases9.
In mammals, there are five SUMO paralogues, SUMO-1 to SUMO-5. To date, no experimental evidence is available about the existence of endogenous SUMO-4 and endogenous SUMO-5 conjugation reactions at the protein level10,11,12. SUMOylation in mammals is carried out by an enzymatic thiol-ester cascade involving three enzymes, the heterodimeric SUMO activating enzyme (SAE1/SAE2) or E1, the SUMO conjugating enzyme (Ubc9) or E2 and a SUMO-E3-ligase specific for each target protein. The action of several families of SUMO E3s appears to be in a dynamic equilibrium with SUMO-specific proteases (SUSPs or SENPs)13 making the SUMOylation reaction highly reversible. Moreover, only a small fraction of the SUMOylated protein versus non-SUMOylated total protein is present. Thereby, isolating endogenous SUMOylated proteins in vivo is rather challenging13.
SUMOylation in vivo was initially studied by western blot using antibodies against the protein of interest14. Immunoprecipitation of the protein was performed with specific antibodies and then PAGE-western blot was carried out with anti-SUMO antibodies. The main problem with this strategy is that antibodies generated against a non-modified recombinant protein are not always able to immunoprecipitate the SUMOylated form of a protein. Alternatively, nickel chromatography after the transient expression of histidine tagged (His6) versions of SUMO molecules and the protein of interest has been used to study SUMOylation in cells. On this basis, it will be more convenient to detect SUMO-modified forms from cells stably expressing His6-SUMO15. For in vivo studies, tandem-SUMO-interacting motifs (SIM) based enrichment was demonstrated for the purification of polySUMO conjugates16. Other groups have been using epitope-tagged antibody SUMO approaches providing a feasible tool to investigate endogenous SUMOylation in primary cells, tissues, and organs17,18. And more recently, Nielsen and colleagues have used antibody-based enrichment to identify endogenous and site-specific SUMO in cells and tissues19.
In order to provide complementary information on the role of SUMOylation in vivo, SUMO-binding entities (SUBEs), also known as SUMO traps, were developed20. Of relevance, tandem ubiquitin binding entities (TUBEs) are considered the conceptual precursors of SUBEs and are commercially available tools for the detection and isolation of polyubiquitylated proteins21. SUBEs are recombinant proteins that comprise tandem repeats of SIMs thereby recognizing SUMO molecules on modified proteins with an increase in the overall affinity for SUMO substrates. SUMO-traps were engineered by introducing an E3 ubiquitin-protein ligase RNF4-derived SIM2 and SIM3 motifs in tandem, into a vector containing glutathione S-transferase (GST), a heterologous carrier protein20. Although SUBEs cannot be used properly to identify mono-SUMOylated target proteins, this method provides a tool to facilitate the purification and identification of poly-SUMO target proteins in vivo. Herein, we describe the application of SUBEs to isolate SUMOylated proteins both in human hepatoma cells and in mouse liver biopsies, an important tool for the study of liver cancer. An overall scheme of the protocol described in this manuscript is shown in Figure 1.
Protocol
All experiments were approved by the CIC bioGUNE institutional committees for animal care and handling. All efforts were made to minimize animal suffering and to reduce the number of animals used. 3-months old male glycine N-methyltransferase (Gnmt) deficient (Gnmt−/−) and its wild type littermates (Gnmt+/+) were used.
1. Cell Preparation and Lysis
NOTE: Herein, Huh-7 (human hepatoma cell line) and THLE2 (human hepatic cell line) were used.
- Maintain cells in P100 plates in standard growth media at 37 °C in a humidified atmosphere of 5% CO2-95% humidity.
- Grow cells in P100 plates plating at a density of 1.2–1.5 × 106 cells per dish by counting the cells using a Neubauer haemocytometry counting chamber.
- Culture Huh-7 in DMEM supplemented with 10% fetal bovine serum (FBS), 1% penicillin-streptomycin-amphotericin B (PSA) and 1% glutamine.
- Culture THLE2 cells in culture dishes pre-coated with 0.01 mg/mL fibronectin, 0.01 mg/mL bovine serum albumin (BSA) and 0.03 mg/mL collagen type I dissolved in growth medium which consists of bronchial epithelial cell growth basal medium (BEGM) supplemented with growth factors (0.4% BPE, 0.1% insulin, 0.1% hydrocortisone, 0.1% retinoic acid, 0.1% transferrin, 0.1% triiodothyronine as well as 10% FBS, 1% PSA, 5 ng/mL epidermal growth factor (EGF) and 70 ng/mL phosphoethanolamine.
- At the end point of the experiment, aspirate the media from the plates and wash cells with 5 mL of sterile 1x phosphate-buffered saline (PBS). Lyse cells directly on the plate placed on ice using 500 µL of lysis buffer (50 mM Tris pH 8.5, 150 mM NaCl, 5 mM EDTA, 1% nonidet P-40 (NP40), supplemented with complete EDTA-free protease inhibitor cocktail and 50 μM PR-619 for each P100 dish. Using a cell scraper, gently scrape the cells off the bottom of the plate into the Lysis medium.
NOTE: Check that all the cells have detached from the plate by visually inspecting the plate base after the treatment. - Alternatively, harvest cells by trypsinization by aspirating cell media and add 1 mL of 1x (0.05%) trypsin-EDTA to the plate, enough to cover the cells and placing the plate in the incubator set at 37 °C, 5% CO2, and 95% humidity for ~5 min ensuring all cells have detached from the plate. Add 2 mL of pre-warmed growth medium in order to stop trypsinization. Centrifuge at 150 g for 10 min and aspirate the supernatant. Wash with 1x PBS and centrifuge at 150 x g for 10 min. After aspirating the supernatant, add 500 µL of lysis buffer (50 mM Tris pH 8.5, 150 mM NaCl, 5 mM EDTA, 1% NP40, supplemented with complete EDTA-free protease inhibitor cocktail and 50 μM PR-619 for each P100 dish.
NOTE: The addition of the PR-619 is critical. - Centrifuge at 15,500 x g and 4°C for 10 min. Transfer the supernatant to another tube and discard the pellet.
NOTE: The protocol can be paused here, and samples stored at -80 °C until further analysis.
2. Tissue Preparation and Lysis
- Upon animal sacrifice, collect mouse livers, wash with cold PBS, and snap freeze immediately in liquid nitrogen. Store the samples −80 °C until further analysis.
- Homogenize 75 mg fragments from snap-frozen/or fresh livers in 1 mL of ice-cold lysis buffer (50 mM Tris pH 8.5, 150 mM NaCl, 5 mM EDTA, 1% NP40, supplemented with complete EDTA-free protease inhibitor cocktail and 50 μM PR-619). Run the homogenizer at 6500 x rpm, 2 x 60 s each, with a 30 s pause (see Table of Materials).
- Centrifuge the samples in a microfuge at 15, 500 x g and 4 °C for 10 min. Transfer the supernatant to another tube and discard the pellet.
- Alternatively, triturate 75 mg of frozen tissues in liquid nitrogen. Then recover the tissue in 1 mL of lysis buffer.
- Centrifuge the sample in a microfuge at 15, 500 x g and 4 °C for 10 min. Transfer the supernatant to another tube and discard the pellet.
NOTE: The protocol can be paused here, and samples stored at -80 °C until further analysis.
3. Binding of GST-SUBEs or GST Control to Glutathione-Agarose Beads
NOTE: The synthesis of the GST-SUBEs or GST control are outside the scope of this manuscript and can be reviewed in previously published literature20. Alternatively, GST- and Control SUBEs are commercially available (e.g., SignalChem).
- Preparation of glutathione beads
- Add 1 mL of de-ionized water to 70 mg of lyophilized glutathione-agarose beads. Reconstitute the beads overnight at 4 °C (or for at least 30 min at room temperature).
- Wash the beads thoroughly after swelling (to remove lactose and ethanol that are usually present in the lyophilized powder agarose beads). To do so, first wash with 10 mL of de-ionized water or PBS followed by centrifugation at 300 x g for 5 min at room temperature. Perform this thrice.
- After 3 washes, resuspend the beads in 1 mL of PBS to obtain a 50% (v/v) slurry.
NOTE: This volume is suitable for the analysis of 10 samples.
- For each sample, add 100 μg of GST-SUBEs or GST control (see reference20) to 100 μL of the glutathione beads slurry and 500 µL of PBS.
NOTE: The relative abundance of the SUMOylated proteins of interest determines the amount of GST-SUBEs used for pull-downs. For each new experimental model, analyze the condition prior to the actual experiment by western blotting the input, bound, and flow-through (FT) material using anti-SUMO2/3 antibodies or against proteins of interest (Liver Kinase B1 (LKB1). - Incubate all the GST-SUBEs or GST control with beads prepared in 3.2., slowly rotating in rotator or mini roller (see Table of Materials) at 4 °C for at least 2 h (slow binding reaction).
NOTE: Adding 1 mM dithiothreitol (DTT) improves GST binding to the glutathione beads. - Recover the agarose beads by centrifugation at 300 x g for 5 min at room temperature. At the end, resuspended the beads in PBS to obtain 50% (v/v) slurry.
NOTE: The protocol can be paused here, and samples stored at -80 ˚C until further analysis.
4. GST Pull Down Assay
- After step 1.5, 2.3 or 2.5, take 1/10 of total volume (e.g. 50 μL) and dilute in the same volume of 3x boiling buffer (250 mM Tris-HCl pH 6.8, 500 mM β-mercaptoethanol, 50% glycerol, 10% SDS, bromophenol blue). This fraction is considered as INPUT.
- Add 450 μL of clarified lysate from steps 1.5, 2.3 or 2.5 to 100 μL glutathione beads slurry. Incubate the lysate with beads, slowly rotating at 4 °C for at least 2 h.
NOTE: Alternatively, 100-200 μg of total protein from steps 1.5, 2.3 or 2.5 (quantified with the Bradford assay) in a total volume of 450 μL can be used. - Spin down the beads in a microfuge at 300 x g for 5 min and collect the supernatant for the analysis. Transfer 1/10 of the total volume (e.g., 50 μL) in a separate tube and dilute in an equal volume of 3x boiling buffer. This fraction is the flow-through (FT) fraction.
- Wash the remaining sample three times with 1 mL ice-cold PBS, 0.05% Tween 20, spin down at 4 °C and 300 x g for 1 min. Carefully aspirate ensuring no liquid remains. The beads correspond to SUBEs BOUND (SB) fraction.
- Elute the sample with 15 μL of 3x boiling buffer and 15 μL of the lysis buffer. This is called the BOUND Fraction.
5. Identification and Characterization of SUMO Targets by Western Blot analysis
- Perform western blot analysis using an anti-SUMO2/3 antibody or any other specific antibody of choice as described previously22.
6. Identification and Characterization of the SUMOylated Proteome by Mass Spectrometry
NOTE: In the case of Mass-Spectrometry (MS) analysis, samples were processed using the Filter-Aided Sample Preparation (FASP) method described by Wisniewski et al.23.
- Desalt the peptides by using stage-tip C18 microcolumns and resuspend them in 0.1% formic acid (FA) prior to MS analysis.
- Load the samples onto a LC-MS system (see Table of Materials) and analyze them in triplicate (technical replicates) (Figure 2b).
- Carry on with the protein identification and abundance calculation by using an associated software.
- For statistical analysis and heatmap generation, load the data onto the Perseus platform (http://www.perseus.tufts.edu/hopper/). Apply a permutation-based false discovery rate (FDR)-corrected t-test for the comparison of the abundances. Proteins with a q < 0.05 and a SUBEs/GST ratio greater than 2 were considered as enriched24.
NOTE: Proteins identified with at least two different peptides are considered in the final analysis.
Representative Results
Identification of a Specific SUMOylation Substrate in Liver Tumor Biopsies by Western Blot Analysis
Liver Kinase B1 (LKB1) SUMOylation has been recently shown to be an important oncogenic driver in liver cancer9,25. Mice deficient in glycine N-methyltransferase (Gnmt), often referred to as Gnmt−/−, is a model that spontaneously develops hepatocellular carcinoma (HCC), the most common type of primary liver cancer. SUBEs were used to enrich and isolate the SUMOylated proteins both in Gnmt−/− mice with liver cancer mice and its wild type littermates (Gnmt+/+). In Figure 2a, Ponceau S staining of the three different fractions (input, FT and BOUND) obtained in the SUBEs pull-down assay are included. A Ponceau S stain is useful to control a possible deleterious effect on the loading of blotted proteins to be evaluated by Western blots. In Figure 2b a Western blot analysis of LKB1 by using SUBEs to capture endogenous SUMOylated LKB1 is shown. The levels of LKB1 SUMOylation are augmented in liver tumors. In the case of Western blot analysis, equal loads and transferred proteins were observed by Ponceau staining of the input fraction and were not significantly altered after washes (flow through fraction). The amount of protein captured with SUBEs was significantly higher, particularly in the tumors. Alternatively, staining a duplicate gel with Coomassie blue can provide similar information. Sticky proteins such as p53 or SUMOylated forms of some proteins might bind to the GST control. To remove the background, use low-density agarose beads, perform a coating with BSA, or incorporate additional washes. However, this could affect applications such as Mass Spectrometry and might result in loss of low affinity interacting proteins.
Characterization of the SUMO Interactome in Human Hepatoma Cells by Mass Spectrometry Analysis
To investigate the capacity of the SUMO-trap to interact with naturally SUMOylated proteins, Huh-7 (human hepatoma) and non-transformed liver epithelial human THLE2 cell lines were used. The first step is the visualization of the total material captured with SUBEs and using GST as a negative control. For this purpose, we can use conventional protein staining protocols as shown in Figure 3a. Then, we performed mass spectrometry analysis. An average of 2268 proteins were identified in the Huh7 GST samples (2339, 2297 and 2168 for each load, respectively), whereas 2812 proteins were identified on average in the Huh7 SUBEs sample (2815, 2817 and 2806). After the subtraction, 742 proteins were enriched in the SUBEs. On the other hand, an average of 2497 proteins were identified in the THLE2 GST samples (2476, 2520 and 2495, respectively) and 2763 in the SUBEs (2823, 2783 and 2684). Of these, 577 were considered to be enriched in the SUBEs samples. Analysis of technical replicates retrieves the heatmap shown in Figure 3b, which was calculated using the default settings available (Euclidean distance, average linkage and pre-processed with k-means). The heatmap depicts the distribution of the 100 most significantly and exclusively enriched proteins in each cell line.

Figure 1: Schematic diagram of the protocol flow chart used for the enrichment, isolation and identification and characterization of the SUMOylated proteome in vivo for the study of liver cancer. Please click here to view a larger version of this figure.
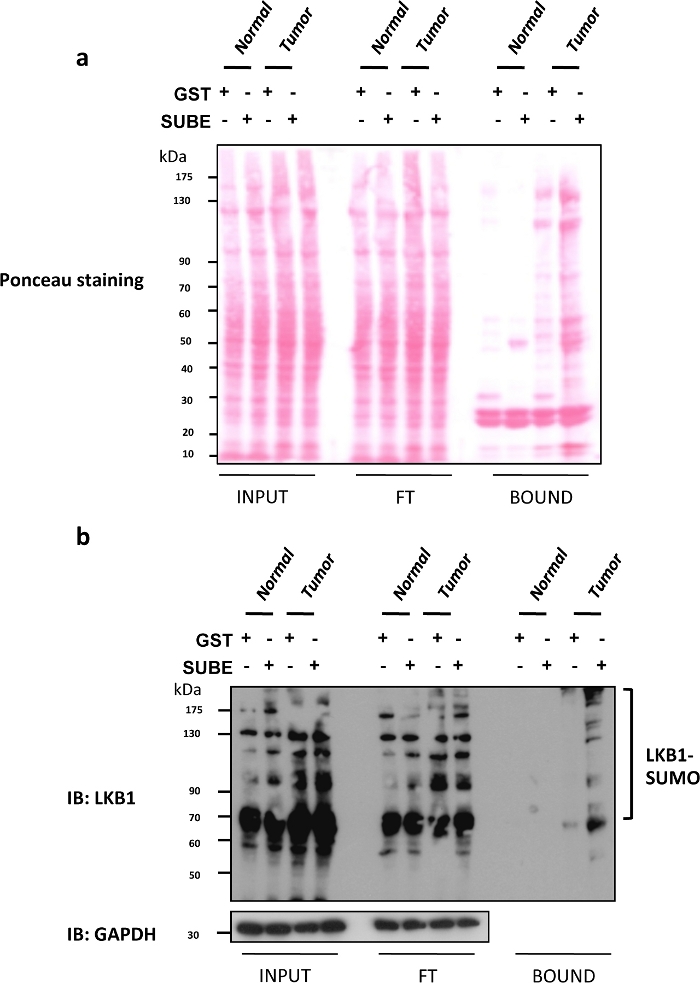
Figure 2: Modification of LKB1 by SUMO-2 in mouse models of Hepatocellular Carcinoma.
(a) Ponceau S staining of the three different fractions (input, Flow through (FT) and BOUND) obtained in the SUBEs pull-down assay. (b) Western blot analysis of LKB1 by using SUMO binding entities (SUBEs) to capture endogenous SUMOylated LKB1; GAPDH is used as a loading control. Please click here to view a larger version of this figure.

Figure 3: Differences between the SUMOylated proteome between the tumoral Huh-7 and non-transformed liver epithelial THLE2 human cell lines.
(a) Sypro staining of captured protein material, with GST (Negative control) and SUBEs. (c) Heatmap depicting the differentially enriched proteins in Huh-7 and THLE2 SUBE samples. Please click here to view a larger version of this figure.
Discussion
Herein, we have provided a complete and detailed description of the methodology reporting the use of SUBEs for the enrichment, isolation and identification and characterization of the SUMOylated proteins in in vivo models of liver cancer. Both in mouse liver tumors and human hepatoma cells, we were able to correctly isolate and identify SUMOylated proteins of interest and to perform a high-throughput characterization of the SUMOylated proteome and interactome. Even though the synthesis of SUBEs is outside the scope of this manuscript, for further information the following references should be looked at26. The protocol described is fast and very sensitive and the critical step of the protocol includes the use of SENPs inhibitors (PR-619). In alternative, chemical isopeptidase inhibitors such as NEM (N-Ethylmaleimide) and IAA (2-Iodoacetamide) in the lysis buffer can be used, however, previous reports have shown that for the SUBEs protocol, the use of PR-619 is advantageous as the other inhibitors interfere with GST binding to the glutathione beads20.
SUBEs are recombinant proteins that comprise tandem repeats of SIMs thereby recognizing SUMO molecules on modified proteins with an increase in the overall affinity for SUMO substrates. Due to its high specificity and sensitivity, the use of SUBEs for the isolation of the SUMOylated proteome is advantageous relative to other approaches in the literature such as the detection by western-blot of specific SUMOylated proteins using antibodies against the protein of interest or the nickel chromatography using the different histidine-tagged versions of SUMO molecules. However, it must be taken into consideration that as the SUBEs protocol is performed under non-denaturing conditions, the interaction between SUMOylated proteins and other interacting proteins is maintained. Therefore, we obtain information about the SUMO interactome rather than only a list of SUMOylated target proteins. Thus, further experiments are necessary to confirm if the identified protein is a SUMO target or an interacting factor. Other limitation of the SUBEs is the fact that control GST traps used are able to capture many background proteins related to oxidative stress. This issue is especially relevant during the MS analysis due to the high sensitivity of the technique. In order to overcome these limitations, biotinylated SUMO-traps (bioSUBEs) have been developed26. Another limitation of SUBEs resides in the fact that we are only able to capture proteins modified by SUMO 2 and SUMO 3 whereas SUMO 1-modified proteins cannot be isolated.
Other concern of the use of SUBEs is related to the amount of starting material necessary for the procedure. The starting material used to capture SUMOylated proteins should consider the different experimental conditions explored. While basal SUMOylation has been reported in various cellular contexts, SUMOylation is a process that is strongly induced after multiple stress conditions/stimuli. If comparing untreated versus treated samples, one has to be sure that the column is not saturated, and differences can be observed between those conditions. In the case of the mouse phenotypes we are analyzing, no treatments have been used and basal SUMOylation levels are low. For this reason, high amounts of proteins were used. The background level should be controlled by using GST and if the unspecific binding is high, the amount of starting material or the binding time should be reduced. The analysis of the FT fraction can be indicative of the capture efficiency even if these traps prefer poly-SUMOylated proteins and a total depletion should not be expected, a reduction of total SUMOylation is in general well observed when the capture efficiency is optimal.
Finally, other application of the SUBEs technology includes the combination of SUBEs technology with Real-time Surface Plasmon Resonance (SPR) allowing the real-time interactions with SUMOylated proteins from cell extracts27. Also, more recently, biotinylated SUMO-traps (bioSUBEs) have been developed with the advantage to reduce the background associated to bigger tags, e.g., during mass spectrometry analysis26. In addition, the bioSUBE version can be used to detect SUMOylated proteins in living cells by fluorescence by using streptavidin-labelled with distinct fluorescent dyes taking advantage of the streptavidin binding to biotin. Also, methods for detection and quantification of SUMOylated proteins can be considered with both GST and bioSUBEs versions such as it was done with the tandem ubiquitin binding entities (TUBEs)21.
Overall, the use of SUBEs for the isolation and characterization of the SUMOylated proteome relevant in liver cancer is a fast and sensitive method providing vast information on the still rather unknown role of the SUMOylation pathway in liver cancer.
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants from Institut National du Cancer, FRANCE, INCa grant PLBIO16-251 (PLBIO16-251), CONACyT-SRE (Mexico) grant 0280365 and the REPERE program of Occitanie, France (M.S.R.). Also, NIH (US Department of Health and Human services)-R01AR001576-11A1, Gobierno Vasco-Departamento de Salud 2013111114 (to M.L.M.-C), ELKARTEK 2016, Departamento de Industria del Gobierno Vasco, MINECO: SAF2017-87301-R integrado en el Plan Estatal de Investigación Cientifica y Técnica y Innovación 2013-2016 cofinanciado con Fondos FEDER, BIOEF (Basque Foundation for Innovation and Health Research): EITB Maratoia BIO15/CA/014; Instituto de Salud Carlos III:PIE14/00031, integrado en el Plan Estatal de Investigación Cientifica y Técnica y Innovación 2013-2016 cofinanciado con Fondos FEDER (to M.L.M.-C), Asociación Española contra el Cáncer (T.C.D, M.L.M-C), Daniel Alagille award from EASL (to T.C.D), Fundación Científica de la Asociación Española Contra el Cancer (AECC Scientific Foundation) Rare Tumor Calls 2017 (to M.L.M), La Caixa Foundation Program (to M.L.M). We thank MINECO for the Severo Ochoa Excellence Accreditation to CIC bioGUNE (SEV-2016-0644).
Materials
| (Gnmt−/−)/ (Gnmt+/+) mice | CIC bioGUNE | ||
| 0.5% Trypsin-EDTA | Life Technologies | 15400-054 | |
| BEBM | Lonza/Clonetics Corporation | cc-3171 | |
| BEGM Bullet Kit | Lonza/Clonetics Corporation | CC3170 | |
| Bromophenol blue | Sigma | 115-39-9 | |
| BSA | Sigma | A4503 | |
| C18 microcolumns | Millipore | Z720070 | |
| Collagen type I | Santa Cruz Biotechnology | sc-136157 | |
| Complete tablets EDTA-free | Roche | 4693132001 | |
| DMEM | Life Technologies | A14431-01 | |
| DTT | Sigma | 43815 | |
| EDTA | Sigma | E6758 | |
| EGF | Sigma | e9644 | |
| FBS | Life Technologies | 10270 | |
| Fibronectin | Life Technologies | 33010018 | |
| Glutamine | Life Technologies | 25030-024 | |
| Glutathione agarose beads | Sigma | G4510 | |
| Glycerol | Sigma | G5516 | |
| GST-Control | SignalChem | G52-30H | |
| GST-SUBEs | SignalChem | S291-340G | |
| Huh7 | CLS (Cell Lines Service) | 300156 | https://clsgmbh.de/ |
| IAA (2-Iodoacetamide) | Merck | L58046844 | |
| LKB1 antibody | Santa Cruz Biotechnology | sc-32245 | |
| Mini LabRoller Rotator | LABNET | H5500 | https://www.labnetinternational.com |
| NaCl | Merck | 106404041000 | |
| nanoElute | BRUKER | https://www.bruker.com/ | |
| NEM (N-Ethylmaleimide) | Sigma | E3876 | |
| NP40 | Fluka | 74385 | |
| PBS | Life Technologies | 14190-094 | |
| Peaks software | Bioinformatics Solutions Inc. | http://www.bioinfor.com/ | |
| Phosphoetanolamine | Sigma | P0503 | |
| Ponceau S solution | Sigma | P7170 | |
| PR-619 | Merck | 662141 | |
| Precellys 24 | Bertin Technologies | P000669-PR240-A | |
| PSA | Life Technologies | 151-40-122 | |
| PSG | Life Technologies | 10378-016 | |
| SDS | Sigma | L3771 | |
| SUMO2/3 antibody | Abcam | Ab3742 | |
| THLE-2 | ATCC | ATCC CRL-2706 | http://www.lgcstandards-atcc.org |
| timsTOF Pro with PASEF mass spectrometer | BRUKER | https://www.bruker.com/ | |
| β-mercaptoethanol | Sigma | 60-24-2 |
References
- Forner, A., Llovet, J. M., Bruix, J. Hepatocellular carcinoma. Lancet. 379 (9822), 1245-1255 (2012).
- Gerbes, A., et al. Gut roundtable meeting paper: selected recent advances in hepatocellular carcinoma. Gut. , (2017).
- Avila, M. A., Berasain, C., Sangro, B., Prieto, J. New therapies for hepatocellular carcinoma. Oncogene. 25 (27), 3866-3884 (2006).
- Grotenbreg, G., Ploegh, H. Chemical biology: dressed-up proteins. Nature. 446 (7139), 993-995 (2007).
- Hendriks, I. A., Vertegaal, A. C. A comprehensive compilation of SUMO proteomics. Nature Reviews in Molecular Cell Biology. 17 (9), 581-595 (2016).
- Seeler, J. S., Dejean, A. SUMO and the robustness of cancer. Nature Reviews Cancer. 17 (3), 184-197 (2017).
- Tomasi, M. L., et al. S-adenosyl methionine regulates ubiquitin-conjugating enzyme 9 protein expression and sumoylation in murine liver and human cancers. Hepatology. 56 (3), 982-993 (2012).
- Li, J., et al. Cbx4 governs HIF-1alpha to potentiate angiogenesis of hepatocellular carcinoma by its SUMO E3 ligase activity. Cancer Cell. 25 (1), 118-131 (2014).
- Zubiete-Franco, I., et al. SUMOylation regulates LKB1 localization and its oncogenic activity in liver cancer. EBioMedicine. 40, 406-421 (2019).
- Sarge, K. D., Park-Sarge, O. K. SUMO and its role in human diseases. Internationa Review of Cell and Molecular Biology. 288, 167-183 (2011).
- Da Silva-Ferrada, E., Lopitz-Otsoa, F., Lang, V., Rodriguez, M. S., Matthiesen, R. Strategies to Identify Recognition Signals and Targets of SUMOylation. Biochemical Research International. , 875148 (2012).
- Liang, Y. C., et al. SUMO5, a Novel Poly-SUMO Isoform, Regulates PML Nuclear Bodies. Science Reports. 6, 26509 (2016).
- Mikolajczyk, J., et al. Small ubiquitin-related modifier (SUMO)-specific proteases: profiling the specificities and activities of human SENPs. Journal of Biologucal Chemistry. 282 (36), 26217-26224 (2007).
- Hilgarth, R. S., Sarge, K. D. Detection of sumoylated proteins. Methods in Molecular Biology. 301, 329-338 (2005).
- Vertegaal, A. C., et al. A proteomic study of SUMO-2 target proteins. Journal of Biological Chemistry. 279 (32), 33791-33798 (2004).
- Bruderer, R., et al. Purification and identification of endogenous polySUMO conjugates. EMBO Reports. 12 (2), 142-148 (2011).
- Becker, J., et al. Detecting endogenous SUMO targets in mammalian cells and tissues. Nature Structural Molecular Biology. 20 (4), 525-531 (2013).
- Barysch, S. V., Dittner, C., Flotho, A., Becker, J., Melchior, F. Identification and analysis of endogenous SUMO1 and SUMO2/3 targets in mammalian cells and tissues using monoclonal antibodies. Nature Protocols. 9 (4), 896-909 (2014).
- Hendriks, I. A., et al. Site-specific characterization of endogenous SUMOylation across species and organs. Nature Communications. 9 (1), 2456 (2018).
- Da Silva-Ferrada, E., et al. Analysis of SUMOylated proteins using SUMO-traps. Science Reports. 3, 1690 (2013).
- Hjerpe, R., et al. Efficient protection and isolation of ubiquitylated proteins using tandem ubiquitin-binding entities. EMBO Reports. 10 (11), 1250-1258 (2009).
- Embade, N., et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology. 55 (4), 1237-1248 (2012).
- Wisniewski, J. R., Zougman, A., Nagaraj, N., Mann, M. Universal sample preparation method for proteome analysis. Nature Methods. 6 (5), 359-362 (2009).
- Meier, F., et al. Online Parallel Accumulation-Serial Fragmentation (PASEF) with a Novel Trapped Ion Mobility Mass Spectrometer. Molecular and Cellular Proteomics. 17 (12), 2534-2545 (2018).
- Ritho, J., Arold, S. T., Yeh, E. T. A Critical SUMO1 Modification of LKB1 Regulates AMPK Activity during Energy Stress. Cell Reports. 12 (5), 734-742 (2015).
- Lang, V., Da Silva-Ferrada, E., Barrio, R., Sutherland, J. D., Rodriguez, M. S. Using Biotinylated SUMO-Traps to Analyze SUMOylated Proteins. Methods in Molecular Biology. 1475, 109-121 (2016).
- Xolalpa, W., Rodriguez, M. S., England, P. Real-Time Surface Plasmon Resonance (SPR) for the Analysis of Interactions Between SUMO Traps and Mono- or PolySUMO Moieties. Methods in Molecular Biology. 1475, 99-107 (2016).

