Carrier-assisted One-pot Sample Preparation for Targeted Proteomics Analysis of Small Numbers of Human Cells
Summary
A protein carrier-assisted one-pot sample preparation coupled with liquid chromatography (LC) – selected reaction monitoring (SRM) termed cLC-SRM is a convenient method for multiplexed targeted proteomics analysis of small numbers of cells, including single cells. It capitalizes on using excessive exogenous protein as a carrier and high-specificity LC-SRM for targeted quantification.
Abstract
Protein analysis of small numbers of human cells is primarily achieved by targeted proteomics with antibody-based immunoassays, which have inherent limitations (e.g., low multiplex and unavailability of antibodies for new proteins). Mass spectrometry (MS)-based targeted proteomics has emerged as an alternative because it is antibody-free, high multiplex, and has high specificity and quantitation accuracy. Recent advances in MS instrumentation make MS-based targeted proteomics possible for multiplexed quantification of highly abundant proteins in single cells. However, there is a technical challenge for effective processing of single cells with minimal sample loss for MS analysis. To address this issue, we have recently developed a convenient protein carrier-assisted one-pot sample preparation coupled with liquid chromatography (LC) – selected reaction monitoring (SRM) termed cLC-SRM for targeted proteomics analysis of small numbers of human cells. This method capitalizes on using the combined excessive exogenous protein as a carrier and low-volume one-pot processing to greatly reduce surface adsorption losses as well as high-specificity LC-SRM to effectively address the increased dynamic concentration range due to the addition of exogeneous carrier protein. Its utility has been demonstrated by accurate quantification of most moderately abundant proteins in small numbers of cells (e.g., 10-100 cells) and highly abundant proteins in single cells. The easy-to-implement features and no need for specific devices make this method readily accessible to most proteomics laboratories. Herein we have provided a detailed protocol for cLC-SRM analysis of small numbers of human cells including cell sorting, cell lysis and digestion, LC-SRM analysis, and data analysis. Further improvements in detection sensitivity and sample throughput are needed towards targeted single-cell proteomics analysis. We anticipate that cLC-SRM will be broadly applied to biomedical research and systems biology with the potential of facilitating precision medicine.
Introduction
Recent technological advances in genomics (transcriptomics) allow for comprehensive and precise analysis of the genome (transcriptome) in single cells1,2,3. However, single-cell proteomics technologies are lagging far behind but are just as important as genomics (transcriptomics) technologies4,5,6,7,8. Furthermore, protein abundance cannot necessarily be inferred from mRNA abundance9, and the proteome is more complex and dynamic than the transcriptome10. Given these challenges, a large number of mixed populations of cells (i.e., bulk cells) are generally used to generate comprehensive proteome data11,12,13. However, such bulk measurements average out stochastic variations of individual cells, thus obscure important cell-to-cell variability (i.e., cell heterogeneity)4,14. Limitations of such bulk measurements become even more severe when the cells of interest only account for a small portion of the total populations of cells (e.g., cancer stem cells within tumors at an early-stage cancer). Therefore, there is a huge knowledge gap between single-cell proteomics and genomics (transcriptomics).
Antibody-based immunoassays (e.g., flow or mass cytometry) are predominantly used for targeted proteomic analysis of single cells6,7,15,16,17,18. However, they suffer from low multiplex, limited specificity, and unavailability of antibodies for new proteins of interest. Mass spectrometry (MS)-based targeted proteomics has emerged as an alternative for accurate protein quantification because of its being antibody-free, high multiplex (≥150 proteins in a single analysis19), high quantitation accuracy (absolute amounts or concentrations), and high specificity and reproducibility (≤10% CV)20,21,22,23. Recent significant progress in sample preparation has made MS-based single-cell proteomics possible for quantitative analysis of highly abundant proteins from single human cells. However, MS-based single-cell proteomics is still at the early infancy stage. For example, the most advanced MS platform coupled with ultralow-flow RPLC flow rates can only allow label-free MS detection and quantification of ~670-870 proteins out of the total ≥12,000 proteins in single HeLa cells24,25.
Currently, there are six MS-based single-cell proteomics approaches available for analysis of single mammalian cells, in which four are for global proteomics (nanoPOTS: nanowell-based Preparation in One pot for Trace Samples26; iPAD-1: integrated proteome analysis device for single-cell analysis27; OAD (oil-air-droplet) chip-based single cell proteomic analysis28; SCoPE-MS: single cell proteomics by mass spectrometry29) and the other two are for targeted proteomics (cLC-SRM: carrier-assisted liquid chromatography (LC) – selected reaction monitoring (SRM)30; cPRISM-SRM: carrier-assisted high pressure, high-resolution separations with intelligent selection and multiplexing coupled to SRM31). However, all these approaches have technical drawbacks. nanoPOTS, iPAD-1, and OAD downscale sample processing volume to 2-200 nL and are not ready for broad benchtop applications26,27,28. For SCoPE-MS, a TMT (tandem mass tag) carrier is added after single-cell processing, so it cannot effectively prevent surface adsorption losses during sample processing when a single tube is used for single-cell processing29, resulting in low reproducibility with a correlation coefficient of only ~0.2-0.4 between replicates32. For cLC-SRM and cPRISM-SRM, using exogenous proteins as a carrier is more suitable for targeted proteomics because peptides from excessive exogenous proteins are frequently sequenced by MS/MS, which greatly reduces the chance for sequencing low abundant endogenous peptides30,31. Unlike global proteomics for relative quantification, the two targeted proteomics approaches can provide accurate or absolute protein analysis of small numbers of cells with high reproducibility using heavy isotope-labeled internal standards at known concentrations. When compared to cPRISM-SRM that requires prior high-resolution PRISM fractionation, resulting in many fraction samples that need to be analyzed, cLC-SRM has a significant advantage in sample throughput without fractionation and can simultaneously quantify hundreds of proteins in a single analysis but with relatively lower detection sensitivity30. Therefore, cLC-SRM is more accessible and should have broader utilities for accurate multiplexed protein analysis of small numbers of cells as well as mass-limited samples.
Herein we describe a detailed protocol to perform cLC-SRM for convenient targeted proteomics analysis of small numbers of human cells, including single cells. The protocol consists of the following major steps: cell sorting by FACS (fluorescence activated cell sorting), cell lysis and digestion processed in low-volume single polymerase chain reaction (PCR) tubes, LC-SRM data collection, and SRM data analysis using publicly available Skyline software (Figure 1). Its broad utility was demonstrated along with our previously well-established SRM assays by absolute targeted quantification of EGFR/MAPK pathway proteins in 1-100 MCF7 or MCF10A cells and determination of pathway protein copies per cell at a wide dynamic range of concentrations30. We anticipate that with the detailed protocol most proteomics researchers can readily implement cLC-SRM in their laboratories for accurate protein analysis of ultrasmall samples (e.g., rare tumor cells) to meet their project needs.
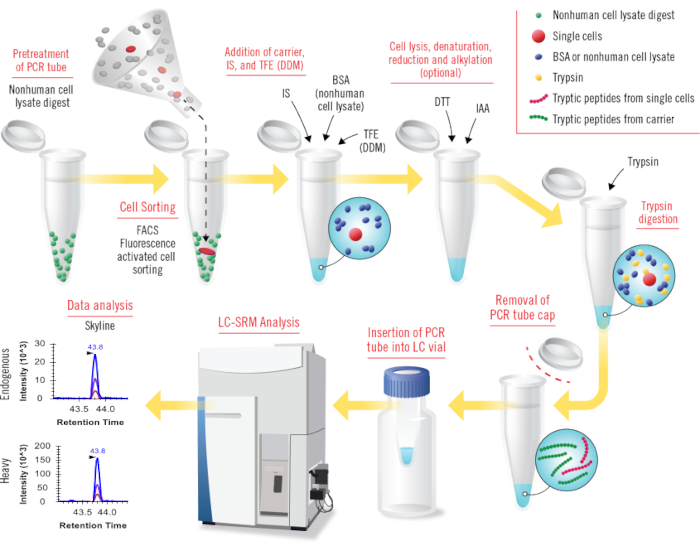
Figure 1: Overview of all steps in cLC-SRM (carried-assisted one-pot sample preparation coupled with liquid chromatography –selected reaction monitoring). Nonhuman cell lysate digests (e.g., Shewanella oneidensis) are used to pretreat PCR tubes for coating tube surface. Small numbers of human cells or single cells sorted by FACS are collected into pretreated PCR tubes. BSA protein (or nonhuman cell lysate proteome) carrier, heavy internal standard (IS), and TFE (or DDM) are added into sample tubes sequentially for facilitating cell lysis and reducing surface adsorption losses. Conceptually, the combined DDM and nonhuman cell lysate proteome carrier will work well for cLC-SRM. Cell lysis is conducted by sonication, and protein denaturation is achieved by heating at high temperature. DTT and IAA reagents are used for reduction and alkylation, respectively (this step is optional). Trypsin is added for digestion with much higher ratios of trypsin over protein amount than that for standard trypsin digestion. The cap of sample tube is removed and then PCR tube is inserted into LC vial for direct LC-SRM analysis. Collected SRM data are analyzed by using publicly available Skyline software. Please click here to view a larger version of this figure.
Protocol
Representative Results
Discussion
cLC-SRM is a convenient targeted proteomics method that enables accurate multiplexed protein analysis of small numbers of cells including single cells. This method capitalizes on protein carrier-assisted one-pot sample preparation, in which all steps including cell collection, multistep cell lysis and digestion, and transfer of peptide digests to capillary LC column for MS analysis are performed in one pot (e.g., single tube or single well) (Figure 1). This 'all-in-one' low-volume on…
Divulgations
The authors have nothing to disclose.
Acknowledgements
This work was supported by NIH Grant R21CA223715 (TS) and UG3CA256967 (TS and HL). The experimental work described herein was performed in the Environmental Molecular Sciences Laboratory, Pacific Northwest National Laboratory, a national scientific user facility sponsored by the United States of America Department of Energy under Contract DE-AC05-76RL0 1830.
Materials
| 2 mL glass LC vial | Microsolv | 9502S-WCV | Vessel to hold PCR tube for autosample injection |
| BSA | Sigma-Aldrich | P0834-10×1mL | Carrier protein for greatly reducing surface adsorption losses |
| DTT | Thermo Scientific | A39255 | Reagent for reduction |
| Formic acid | Thermo Scientific | 28905 | Reagent for stopping enzyme reaction |
| IAA | Thermo Scientific | A39271 | Reagent for alkylation |
| Peptide internal standards | New England peptide | Targeted quantification of EGFR/MAPK pathway proteins | |
| RT-PCR tube | GeneMate Bioexpress | T-3035-1 | 0.2 mL PCR tube for one-pot sample preparation |
| Skyline software | University of Washington | Publicly available for SRM data analysis | |
| Sonicator | Hielscher Ultrasound Technology | UTR200 | Sonication on ice for cell lysis |
| Speed Vac concentrator | Thermo Scientific | Reduction of the percentage of TFE for effective trypsin digestion | |
| TFE | Sigma-Aldrich | 18370-10×1mL | 60% TFE for cell lysis |
| Thermocycler w/ heated lid | Peltier Thermal Cycler | PTC-200 | Heating for protein denaturation |
| Trypsin Gold | Promega | V528A | Enzyme for protein digestion |
| Waters BEH C18 column | Waters | C18 column for peptide separation |
References
- Wang, Z., Gerstein, M., Snyder, M. RNA-Seq: a revolutionary tool for transcriptomics. Nature Reviews Genetics. 10 (1), 57-63 (2009).
- Miyamoto, D. T., et al. RNA-Seq of single prostate CTCs implicates noncanonical Wnt signaling in antiandrogen resistance. Science. 349 (6254), 1351-1356 (2015).
- Wang, Y., Navin, N. E. Advances and applications of single-cell sequencing technologies. Molecular Cell. 58 (4), 598-609 (2015).
- Macaulay, I. C., Ponting, C. P., Voet, T. Single-Cell Multiomics: Multiple Measurements from Single Cells. Trends in Genetics. 33 (2), 155-168 (2017).
- Gavasso, S., Gullaksen, S. E., Skavland, J., Gjertsen, B. T. Single-cell proteomics: potential implications for cancer diagnostics. Expert Review of Molecular Diagnostics. 16 (5), 579-589 (2016).
- Wu, M., Singh, A. K. Single-cell protein analysis. Current Opinion in Biotechnology. 23 (1), 83-88 (2012).
- Hughes, A. J., et al. Single-cell western blotting. Nature Methods. 11 (7), 749-755 (2014).
- Peterson, V. M., et al. Multiplexed quantification of proteins and transcripts in single cells. Nature Biotechnology. 35 (10), 936-939 (2017).
- Liu, Y., Beyer, A., Aebersold, R. On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 165 (3), 535-550 (2016).
- Choudhary, C., Mann, M. Decoding signalling networks by mass spectrometry-based proteomics. Nature Reviews Molecular Cell Biology. 11 (6), 427-439 (2010).
- Mertins, P., et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 534 (7605), 55-62 (2016).
- Zhang, H., et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell. 166 (3), 755-765 (2016).
- Zhang, B., et al. Proteogenomic characterization of human colon and rectal cancer. Nature. 513 (7518), 382-387 (2014).
- Gaudet, S., Miller-Jensen, K. Redefining Signaling Pathways with an Expanding Single-Cell Toolbox. Trends in Biotechnology. 34 (6), 458-469 (2016).
- Giesen, C., et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nature Methods. 11 (4), 417-422 (2014).
- Di Palma, S., Bodenmiller, B. Unraveling cell populations in tumors by single-cell mass cytometry. Current Opinion in Biotechnology. 31, 122-129 (2015).
- Schulz, K. R., Danna, E. A., Krutzik, P. O., Nolan, G. P. Single-cell phospho-protein analysis by flow cytometry. Current Protocols in Immunology. , 11-20 (2012).
- Willison, K. R., Klug, D. R. Quantitative single cell and single molecule proteomics for clinical studies. Current Opinion in Biotechnology. 24 (4), 745-751 (2013).
- Lee, J. Y., et al. Detection of Head and Neck Cancer Based on Longitudinal Changes in Serum Protein Abundance. Cancer Epidemiology, Biomarkers & Prevention. 29 (8), 1665-1672 (2020).
- Shi, T., et al. Advancing the sensitivity of selected reaction monitoring-based targeted quantitative proteomics. Proteomics. 12 (8), 1074-1092 (2012).
- Shi, T., et al. Advances in targeted proteomics and applications to biomedical research. Proteomics. 16 (15-16), 2160-2182 (2016).
- Lange, V., Picotti, P., Domon, B., Aebersold, R. Selected reaction monitoring for quantitative proteomics: a tutorial. Molecular Systems Biology. 4, 222 (2008).
- Picotti, P., Aebersold, R. Selected reaction monitoring-based proteomics: workflows, potential, pitfalls and future directions. Nature Methods. 9 (6), 555-566 (2012).
- Zhu, Y., et al. Proteomic Analysis of Single Mammalian Cells Enabled by Microfluidic Nanodroplet Sample Preparation and Ultrasensitive NanoLC-MS. Angewandte Chemie International Edition. 57 (38), 12370-12374 (2018).
- Cong, Y., et al. Improved Single-Cell Proteome Coverage Using Narrow-Bore Packed NanoLC Columns and Ultrasensitive Mass Spectrometry. Analytical Chemistry. 92 (3), 2665-2671 (2020).
- Zhu, Y., et al. Nanodroplet processing platform for deep and quantitative proteome profiling of 10-100 mammalian cells. Nature Communications. 9 (1), 882 (2018).
- Shao, X., et al. Integrated Proteome Analysis Device for Fast Single-Cell Protein Profiling. Analytical Chemistry. 90 (23), 14003-14010 (2018).
- Li, Z. Y., et al. Nanoliter-Scale Oil-Air-Droplet Chip-Based Single Cell Proteomic Analysis. Analytical Chemistry. 90 (8), 5430-5438 (2018).
- Budnik, B., Levy, E., Harmange, G., Slavov, N. SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biology. 19 (1), 161 (2018).
- Zhang, P., et al. Carrier-Assisted Single-Tube Processing Approach for Targeted Proteomics Analysis of Low Numbers of Mammalian Cells. Analytical Chemistry. 91 (2), 1441-1451 (2019).
- Shi, T., et al. Facile carrier-assisted targeted mass spectrometric approach for proteomic analysis of low numbers of mammalian cells. Communications Biology. 1, 103 (2018).
- Vitrinel, B., Iannitelli, D. E., Mazzoni, E. O., Christiaen, L., Vogel, C. Simple Method to Quantify Protein Abundances from 1000 Cells. ACS Omega. 5 (25), 15537-15546 (2020).
- MacLean, B., et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26 (7), 966-968 (2010).
- Shi, T., et al. Conservation of protein abundance patterns reveals the regulatory architecture of the EGFR-MAPK pathway. Science Signaling. 9 (436), 6 (2016).
- Addona, T. A., et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nature Biotechnology. 27 (7), 633-641 (2009).
- He, J., et al. Analytical platform evaluation for quantification of ERG in prostate cancer using protein and mRNA detection methods. Journal of Translational Medicine. 13, 54 (2015).
- Shi, T., et al. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proceedings of the National Academy of Sciences. 109 (38), 15395-15400 (2012).
- Shi, T., et al. Targeted quantification of low ng/mL level proteins in human serum without immunoaffinity depletion. Journal of Proteome Research. 12 (7), 3353-3361 (2013).

.
