Micropatterned Surfaces to Study Hyaluronic Acid Interactions with Cancer Cells
Summary
A novel approach that allows the high-resolution analysis of cancer cell interactions with exogenous hyaluronic acid (HA) is described. Patterned surfaces are fabricated by combining carbodiimide chemistry and microcontact printing.
Abstract
Cancer invasion and progression involves a motile cell phenotype, which is under complex regulation by growth factors/cytokines and extracellular matrix (ECM) components within the tumor microenvironment. Hyaluronic acid (HA) is one stromal ECM component that is known to facilitate tumor progression by enhancing invasion, growth, and angiogenesis1. Interaction of HA with its cell surface receptor CD44 induces signaling events that promote tumor cell growth, survival, and migration, thereby increasing metastatic spread2-3. HA is an anionic, nonsulfated glycosaminoglycan composed of repeating units of D-glucuronic acid and D-N-acetylglucosamine. Due to the presence of carboxyl and hydroxyl groups on repeating disaccharide units, native HA is largely hydrophilic and amenable to chemical modifications that introduce sulfate groups for photoreative immobilization 4-5. Previous studies involving the immobilizations of HA onto surfaces utilize the bioresistant behavior of HA and its sulfated derivative to control cell adhesion onto surfaces6-7. In these studies cell adhesion preferentially occurs on non-HA patterned regions.
To analyze cellular interactions with exogenous HA, we have developed patterned functionalized surfaces that enable a controllable study and high-resolution visualization of cancer cell interactions with HA. We utilized microcontact printing (uCP) to define discrete patterned regions of HA on glass surfaces. A “tethering” approach that applies carbodiimide linking chemistry to immobilize HA was used 8. Glass surfaces were microcontact printed with an aminosilane and reacted with a HA solution of optimized ratios of EDC and NHS to enable HA immobilization in patterned arrays. Incorporating carbodiimide chemistry with mCP enabled the immobilization of HA to defined regions, creating surfaces suitable for in vitro applications. Both colon cancer cells and breast cancer cells implicitly interacted with the HA micropatterned surfaces. Cancer cell adhesion occurred within 24 hours with proliferation by 48 hours. Using HA micropatterned surfaces, we demonstrated that cancer cell adhesion occurs through the HA receptor CD44. Furthermore, HA patterned surfaces were compatible with scanning electron microscopy (SEM) and allowed high resolution imaging of cancer cell adhesive protrusions and spreading on HA patterns to analyze cancer cell motility on exogenous HA.
Protocol
1. Standard Photolithography for Micropatterned Stamp Fabrication
- Rinse a new silicon wafer with ethanol and dry with stream of air. Use forceps to handle wafer and prevent surface defects during the entire process.
- Transfer wafer to spin coater and cover wafer surface with SU-2025 negative photoresist. Cover at least 80% of wafer with photoresist. Spin coat for 10 seconds at 600rpm, followed by 30 seconds at 3000rpm. Due to the photosensitivity of the photoresist, turn off room lights from this point forward.
- Transfer photoresist coated silicon wafer to hot plate for “soft-bake” at 95°C for 3 minutes. Remove from hot plate and allow 1 minute to cool.
- Place a pre-fabricated mask with the desired pattern on top of the photoresist-covered silicon wafer and expose to ultraviolet (UV) irradiation (350-450 nm) for 20 seconds.
- Transfer silicon wafer to hot plate for “hard-bake” at 110°C for 6 minutes.
- Turn on lights and remove wafer from hot plate. Distinct patterns should be visible at this point.
- Carefully rinse silicon wafer with SU-8 photoresist developer. After 3 rinses with developer solution, rinse with ethanol. If any white residue is present, repeat rinse with photoresist developer solution, or simply submerge the wafer in developer solution for 2 minutes and proceed again with ethanol rinse. Wash with water and air dry.
- Mix polydimethylsiloxane (PDMS) elastomer solution and curing agent in a 10:1 weight ratio. Centrifuge at 900rpm for 5 minutes to remove air bubbles.
- Place patterned wafer in Petri dish and pour PDMS onto silicon wafer. If there are bubbles present, place in desicator and vacuum for a couple minutes until bubbles disappear.
- Cure overnight at room temperature (RT) to form a complementary elastomeric stamp. If PDMS is not completely cured after 12 hours, place in oven at 60°C for 30 minutes.
- Carefully remove PDMS from silicon wafer. Use a razor edge to cut stamp to desired size. Sonicate in ethanol for 15 minutes. Wafers can be utilized multiple times to create new elastomeric stamps.
2. HA Micropatterning
- Plasma clean glass slides for 5 minutes. Place all slides into chamber and vacuum for 5 minutes. Then allow low flow of oxygen to enter chamber. Turn plasma cleaner on for 5 minutes. Remove from plasma cleaner and place in a Petri dish using forceps.
- During plasma cleaning, sonicate PDMS stamps in ethanol for 15 minutes.
- Prepare a fresh 3% v/v solution of 3-aminopropyltrimethoxysilane (APTMS) in 95% v/v ethanol in a 15 mL centrifuge tube. Allow to react for 5 minutes at RT to form a silanol.
- Place PDMS stamp pattern-up on spin coater chuck and cover patterned surface with APTMS solution. Spincoat at 3500rpm for 30 seconds. Do not touch the stamp surface, only the sides of PDMS stamp.
- Transfer APTMS solution to the plasma cleaned glass surfaces: set stamp facing down on one end of patterned side and gently glide pressing PDMS so it is completely in contact with glass surface for 1 minute.
- Remove PDMS stamp gently, and allow patterned glass slide to incubate in RT for 30 minutes. Rinse patterned surfaces with ethanol and air dry. Sonicate PDMS stamp for 15 minutes to remove excess APTMS.
- Heat surfaces in oven or on hot plate for 1 hour at 115°C. Rinse with ethanol and dry with air.
- Prepare fresh polyethylene glycol (PEG)-silane solution of 20% v/v 2-[methoxy(polyethyleneoxy)propyl] trimethoxysilane in toluene. This will serve as a non-adhesive region surrounding patterns.
- Transfer patterned glass slide to a glass Petri dish and incubate in PEG-silane solution for 45 minutes at 75°C on a hot plate. Rinse with toluene, water, and dry with air.
- Sterilize for 1 hour using UV germicidal irradiation in a biological safety cabinet. Proceed in biological safety cabinet from this point forward to maintain sterility.
- Prepare aqueous sterile HA solution.
10mM of 1-Ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC)
5mM of N-hydroxysuccinimide
50μg/ mL fluorescein-labeled HA - Apply HA solution to the patterned glass substrates in sterile conditions and protected from light. Patterned substrates should be undisturbed for 16-24 hours.
- Aspirate off HA solution, wash with phosphate-buffered saline (PBS), and cover surface with 1% bovine serum albumin (BSA) in PBS for 1 hour. The surface is now ready for cell culture.
3. Cell Culture on HA Micropatterned Surfaces
- Expand MDA-MB-231 human breast cancer cells and LS174T colorectal carcinoma cells, according to the manufacturer’s instructions.
- Seed single cell suspensions of cancer cells with densities between 0.4 and 1×106 cancer cells per 18.75 cm2 glass surface area and onto HA immobilized surfaces. Incubate in humidified incubator (37 °C) in atmospheres maintained with 5% CO2. Cell adhesion should occur within 24 hours and can be observed using inverted light microscope
- Cell culture surfaces can be maintained up to 5 days in standard incubator conditions. Change media every other day. Cell morphology and growth can be observed using inverted light microscope. Additional cellular assays can be applied to analyze responses to the HA surface.
4. Representative Results:
The carbodiimide chemistry used to covalently immobilize HA to APTMS patterned glass slides is shown in Figure 1A. EDC is a zero-length crosslinking agent that reacts with HA carboxyl groups to form amine-reactive intermediates. As this intermediate is susceptible to hydrolysis, NHS is added to increase carbodiimide reaction efficiency. As HA solution interacts with APTMS micropatterns, a stable amide bond forms between HA intermediate and primary amine of ATPMS. An example of HA patterned surface visualized using a fluorescent microscope and an ImageJ fluorescence intensity analysis demonstrate the patterned immobilization of HA are shown (Figure 1B-C).
The HA micropatterned surfaces enable the study of cancer cell interactions with expgenous HA. Both MDA-MB-231 human breast and LS174t human colon cancer cell lines preferentially adhere to HA micropatterned regions (Figure 2). High resolution imaging using scanning electron microscopy can be used to analyze interactions of cultured cells with HA immobilized surfaces (Figure 3).
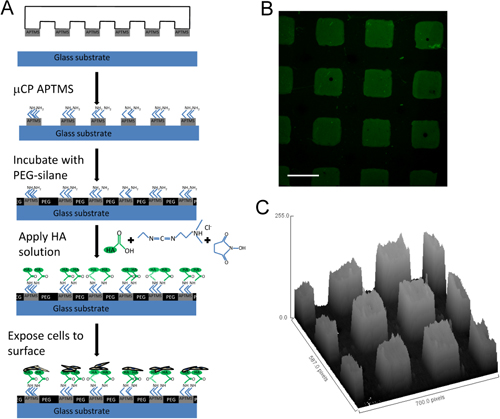
Figure 1 HA micropatterned surfaces (A) Schematic describing the development of HA functional surfaces, (B) fluorescence image of fluorescein (green)-labeled HA micropatterned surface. (C) 3D pixel distribution map using ImageJ software demonstrating the discrete HA patterns. Scale bar = 100μm. Modified from 9 with permission from Elsevier.
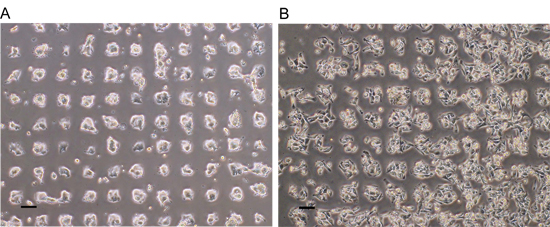
Figure 2. Cancer cell adhesion on HA micropatterned surfaces. Both colon (A) and breast (B) carcinoma preferentially adhered onto HA patches with 24 hours of culture. Modified from 9 with permission from Elsevier.
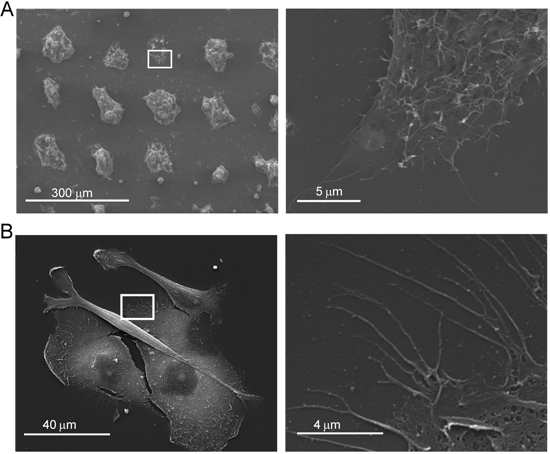
Figure 3. Cancer cell interaction with HA. (A) Scanning electron microscopy images of colon cancer cells (A) cultured on HA surfaces shown at low magnification (left) and high magnification (right; of squares on left) and of breast cancer cells (B) cultured on HA surfaces shown at low magnification (left) and high magnification (right; of squares on left).
Discussion
The HA micropatterning method presented allows the study of cell interactions with exogenous HA. HA is known to play a key role in cancer progression 1 however there have been limited studies investigating the interaction of cancer cells on two dimensional HA patterned surfaces. A controllable study on exogenous HA micropatterns allows high-resolution visualization of cancer cell adhesion, growth, and migration and may elucidate further fundamentals of cancer.
By combining carbodiimide chemistry and microcontact printing, we have developed surfaces with distinct HA patterns to study cancer cell interactions. This method allows consistently patterned surfaces compatible with cell culture and analysis techniques including, but not limited to immunofluorescent staining, scanning electron microscopy, inhibition, migration, etc.
Additionally, photolithographic techniques allow the easy microfabrication of any desired pattern, to control immobilization of HA for specific applications. For example, fabricating PDMS stamps with linear stripes would immobilize HA in a linear array and could be useful in analyzing cell migration along patterned HA.
Although we have primarily utilized this method to elucidate cancer cell interactions with HA, this technique is applicable to study interactions with other cell types. Additionally, this method can be used to immobilize a wide range of HA molecules. The HA we have immobilized is fluorescently labeled with a molecular weight of 800 kDa. However, HA ranges in size from 2000- 25000 repeating disaccharide units, and cellular response has been demonstrated to be dependent on HA molecular weight 10-11. This protocol can therefore be applied to immobilize HA of varying molecular weights to investigate cell response to exogenous HA.
Divulgations
The authors have nothing to disclose.
Acknowledgements
The authors acknowledge the use of the surface analysis laboratory at Johns Hopkins, funded as part of the Materials Research Science and Engineering Center through the National Science Foundation. LED is an IGERT trainee and a National Science Foundation Graduate Fellow. This research was partially supported by NIH grant U54CA143868.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| SU-2025 photoresist | MicroChem Corp. | Y111069 | ||
| SU-8 developer | MicroChem Corp. | Y020100 | ||
| Sylgard 184 | Dow Corning | |||
| 3-aminopropyltrimethoxysilane (APTMS) | Sigma-Aldrich | 281778 | ||
| 2- [methoxy(polyethyleneoxy) propyl] trimethoxysilane (Peg-silane) | Gelest Inc | SIM6492.7 | ||
| 1-Ethyl-3- (3-dimethylaminopropyl) carbodiimide | Thermo Scientific | 22980 | ||
| N-hydroxysuccinimide (NHS) | Thermo Scientific | 24500 | ||
| Fluorescein labeled hyaluronic acid (FL-HA) | Sigma-Aldrick | F1177 | Reconstitute with 10ml of DI water | |
| MDA-MB-231 breast carcinoma cells | ATCC | HTB-26 | ||
| LS174t colon carcinoma cells | ATCC | Cl-188 |
References
- Toole, B. P., Wight, T. N., Tammi, M. I. Hyaluronan-cell interactions in cancer and vascular disease. Journal of Biological Chemistry. 277, 4593-4596 (2002).
- Hamilton, S. R. The Hyaluronan Receptors CD44 and Rhamm (CD168) Form Complexes with ERK1,2 That Sustain High Basal Motility in Breast Cancer Cells. Journal of Biological Chemistry. 282, 16667-16680 (2007).
- Götte, M., Yip, G. W. Heparanase, Hyaluronan, and CD44 in Cancers: A Breast Carcinoma Perspective. Recherche en cancérologie. 66, 10233-10237 (2006).
- Lord, M. S., Pasqui, D., Barbucci, R., Milthorpe, B. K. Protein adsorption on derivatives of hyaluronic acid and subsequent cellular response. Journal of Biomedical Materials Research. Part A 91, 635-646 (2009).
- Barbucci, R. Modification of hyaluronic acid by sulphate groups insertion to obtain a heparin-like molecule. Gazz. Chim. Ital. 125, 169-180 (1995).
- Morra, M., Cassinelli, C. Non-fouling properties of polysaccharide-coated surfaces. Journal of Biomaterials Science, Polymer Edition. 10, 1107-1124 (1999).
- Khademhosseini, A. Layer-by-layer deposition of hyaluronic acid and poly-L-lysine for patterned cell co-cultures. Biomaterials. 25, 3583-3592 (2004).
- Ibrahim, S., Joddar, B., Craps, M., Ramamurthi, A. A surface-tethered model to assess size-specific effects of hyaluronan (HA) on endothelial cells. Biomaterials. 28, 825-835 (2007).
- Dickinson, L. E., Ho, C. C., Wang, G. M., Stebe, K. J., Gerecht, S. Functional surfaces for high-resolution analysis of cancer cell interactions on exogenous hyaluronic acid. Biomaterials. 31, 5472-5478 (2010).
- Gao, F. Hyaluronan oligosaccharides promote excisional wound healing through enhanced angiogenesis. Matrix Biology. 29, 107-116 (2010).
- Slevin, M. Hyaluronan-mediated angiogenesis in vascular disease: Uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biology. 26, 58-68 (2007).

