High-throughput Analysis of Mammalian Olfactory Receptors: Measurement of Receptor Activation via Luciferase Activity
Summary
Olfactory receptor activation patterns encode odor identity, but the lack of published data identifying odorant ligands for mammalian olfactory receptors hinders the development of a comprehensive model of odor coding. This protocol describes a method to facilitate high-throughput identification of olfactory receptor ligands and quantification of receptor activation.
Abstract
Odorants create unique and overlapping patterns of olfactory receptor activation, allowing a family of approximately 1,000 murine and 400 human receptors to recognize thousands of odorants. Odorant ligands have been published for fewer than 6% of human receptors1-11. This lack of data is due in part to difficulties functionally expressing these receptors in heterologous systems. Here, we describe a method for expressing the majority of the olfactory receptor family in Hana3A cells, followed by high-throughput assessment of olfactory receptor activation using a luciferase reporter assay. This assay can be used to (1) screen panels of odorants against panels of olfactory receptors; (2) confirm odorant/receptor interaction via dose response curves; and (3) compare receptor activation levels among receptor variants. In our sample data, 328 olfactory receptors were screened against 26 odorants. Odorant/receptor pairs with varying response scores were selected and tested in dose response. These data indicate that a screen is an effective method to enrich for odorant/receptor pairs that will pass a dose response experiment, i.e. receptors that have a bona fide response to an odorant. Therefore, this high-throughput luciferase assay is an effective method to characterize olfactory receptors—an essential step toward a model of odor coding in the mammalian olfactory system.
Introduction
The mammalian olfactory system has the ability to respond to a vast number of odorous stimuli, allowing for the detection and discrimination of thousands of odorants. Olfactory receptors (ORs) are the molecular sensors expressed by the olfactory sensory neurons in the olfactory epithelium12. Mammalian odor recognition occurs through differential activation of ORs by odorants, and the OR gene family is extensive, with roughly 1,000 murine and 400 human receptors12-16. Previous functional analyses of ORs in olfactory neurons and in heterologous cells have shown that different odorants are recognized by unique, but overlapping ensembles of ORs10,17-20. Matching ligands to ORs is critical for understanding the olfactory code and essential for building viable models of olfaction. Due to difficulties expressing ORs in heterologous systems as well as the large number of both odorants and ORs, this data has been largely absent from the field; indeed, fewer than 6% of human ORs have a published ligand1-11. This protocol describes the use of a luciferase assay to characterize odorant/OR interactions. This assay enables the high-throughput characterization of ORs, a task that is essential to understanding odorant/OR interactions as well as developing a model of odor coding.
High-throughput studies of ORs face three major challenges. First, ORs expressed in heterologous cells were retained in the ER and subsequently degraded in the proteasome21,22, preventing the ORs from interacting with odorants in the assay system23-25. This problem was addressed by the discovery of accessory proteins that facilitate stable cell-surface expression of a broad range of ORs19,26,27. Receptor-transporter-proteins 1 and 2 (RTP1 and 2) promote OR cell-surface expression and activation in response to odorant stimulation19. Based on this work, HEK293T cells were modified to stably express RTP1 long (RTP1L) and RTP2, receptor expression-enhancing protein 1, and Gαolf, resulting in the Hana3A cell line19,27. In addition, the type 3 muscarinic acetylcholine receptor (M3-R) interacts with ORs at the cell surface and enhances activation in response to odorants26. Co-transfection of an OR with RTP1S and M3-R into Hana3A cells results in the robust, consistent, and functional expression of a broad range of ORs at the cell surface27. Second, mammalian OR repertoires are quite large. In humans, for example, the OR repertoire is an order of magnitude more numerous than the gustatory receptor repertoire, and 2 orders of magnitude more numerous than the visual receptor repertoire. Although cloning a single OR is a relatively straightforward protocol, significant up-front effort is required to generate a comprehensive library. Third, although we know that in vision, wavelength translates into color and in audition frequency translates into pitch, the organization of odors is poorly understood, making it difficult for researchers to interpolate from a representative sample of odorants. Although some progress has been made on this front10,28, the map of the olfactory landscape remains incomplete. Screening tens of thousands of molecules against hundreds of ORs is a daunting task; high-throughput screens in this domain require carefully defined campaigns. The major remaining challenges are those of logistics and cost rather than problems inherent to the technique. Although heterologous screening has not been widely used to identify ligands by academic groups, a private company has used the same technique to identify ligands for 100 human ORs 29. Unfortunately, these data remain proprietary.
The high-throughput luciferase assay outlined here has several advantages over alternative methods used to assess OR activation. Although the responses of native olfactory sensory neurons have been measured using electrophysiology and calcium imaging, these techniques have difficulty teasing apart which OR leads to a neuron's response due to the overlap in response properties for olfactory neurons. Although knocking-in a GFP-labeled receptor type30,31, delivering specific receptors via adenovirus to murine olfactory neurons32,33, or performing RT-PCR after recordings17,24,33 can link recordings to single receptor types, these methods are low-throughput and not suitable for large-scale screens. Heterologous screening systems are more scalable, and two major forms are found in the literature: cAMP pathway reporters and inositol triphosphate (IP3) pathway reporters. Upon odor stimulation, ORs activate a Gαolf transduction signaling cascade that results in the production of cyclic AMP (cAMP)12. By co-transfecting a firefly luciferase reporter gene under the control of a cAMP response element (CRE), luciferase production can be measured as a function of odor response, allowing for the quantification of OR activation. OR activation can also be linked to the IP3 pathway by co-expressing G-proteins such as Gα15/16 or a Gα15-olf chimera24,25,34. We have chosen the assay presented here based on three factors: (1) the co-expression of RTP1 with Rho-tagged olfactory receptors improves the expression of olfactory receptors at the cell surface19,27; (2) use of a cAMP-responsive reporter gene allows for the measurement of OR activation through the canonical second messenger pathway; and (3) the assay is well-suited to high-throughput screens.
This high-throughput luciferase assay is applicable to a variety of studies valuable to the field of olfaction. First, a large number of ORs can be screened against a single odorant in order to determine the receptor activation pattern for a specific odorant. This type of study identified OR7D4 as the OR responsible for responding to the steroid odorant androstenone8. Conversely, one OR can be screened against a panel of odorants in order to determine the receptor response profile10. When candidate olfactory odorant/OR pairs are identified via these screens, interaction can be confirmed by conducting a dose response experiment examining the response of the OR to increasing concentrations of odorant. Dose response curves can also assess how genetic variation in an OR affects in vitro odorant response8,9,11,35, and these studies can be extended to interspecific OR variation, allowing for the examination of receptor evolution across species and causal mutations in evolution36,37, Finally, this assay can be used to screen for odor antagonists that are able to antagonize OR response to a particular odorant for a known odorant/receptor pair38,39. In summary, this high-throughput luciferase assay is applicable to a range of studies that will help characterize OR activation patterns and provide a better understanding of odor coding in the olfactory system.
Protocol
1. Culture of Hana3A Cells
- Prepare M10 media by supplementing minimum essential medium (MEM) with 10% (v/v) FBS.
- Culture Maintenance
- Maintain cells in M10 media. NOTE: The expression vectors for RTP1L, RTP2, REEP1, and Gαolf confer puromycin resistance to Hana3A cells, but maintaining the cells with this antibiotic does not significantly affect assay results.
- Subculture at a ratio of 1:8 in 10 cm dishes every 2-3 days.
- Incubate at 37 °C with 5% CO2.
2. Plating Cells for Transfection
- Aspirate media from a 100% confluent 10 cm dish of Hana3A cells.
- Wash cells by adding 10 ml PBS, swirling the dish, and aspirating the PBS.
- Add 3 ml of 0.05% trypsin/EDTA and wait for cells to dissociate (about 1 min).
- Inactivate trypsin by adding 5 ml M10 and break up cell clumps by triturating roughly 10x with a 10 ml pipette. Pipette carefully to avoid introducing air bubbles into the media.
- For each 96-well plate, transfer 1 ml of cells into a 15 ml conical tube, centrifuge at 200 x g for 5 min, and aspirate the supernatant without disturbing the cell pellet.
- Resuspend the cells in 6 ml M10 per 1 ml of cells transferred in Step 2.5.
- Pipette 50 µl of cells to each well of a 96-well plate and incubate overnight at 37 °C with 5% CO2.
3. Transfection of Olfactory Receptors
- Preparation of Plasmid DNA
- Prepare plasmid DNA via an endotoxin-free protocol. NOTE: Use plasmid DNA preparation kits designated "endotoxin-free," or add a phenol-chloroform extraction step to the plasmid DNA preparation protocol.
- Dilute DNA to a concentration of 100 ng/µl in TE buffer.
- Observe plated cells (Step 2.7) to ensure a proper confluency of approximately 30-50% per well and return to incubator. NOTE: While this confluency is not optimal for the lipid transfection reagent, a confluency of 30-50% at this step is optimal for measuring luciferase activity 24 hr after transfection.
- Preparation of Transfection Mix
- Pipette RTP1S-pCI, M3-R-pCI, pCRE-luc, and pSV40-RL plasmids into MEM medium per the volumes detailed in Table 1 to make the Plasmid mix (volumes indicated are per 96-well plate).
Plasmid mix per well per 96-well plate MEM — 500 µl RTP1S-pCI 5 ng 480 ng M3-R-pCI 2.5 ng 240 ng pCRE-luc 10 ng 960 ng pSV40-RL 5 ng 480 ng
Table 1. Plasmid mix components. Per well and per 96-well plate volumes of RTP1S-pCI, M3-R-pCI, pCRE-luc, and pSV40-RL, and MEM. - For each 96-well plate, dilute 18 µl lipid transfection reagent in 450 µl MEM medium.
- Pipette Plasmid mix (from Step 3.3.1), rhodopsin-tagged olfactory receptor in pCI plasmid (Rho-OR-pCI), and lipid transfection mix (from Step 3.3.2) to make the Complex detailed in Table 2. Mix the solution by trituration and incubate at room temperature for 15 min. Stop the reaction by adding M10 according to Table 2. NOTE: this reaction is time-sensitive and should not be allowed to continue for more than 30 min. The well + 10% calculation is important to ensure sufficient volume for subsequent steps.
Complex per well per well + 10% Plasmid mix 4.2 µl 4.58 µl Rho-OR-pCI 0.05 ng 0.06 ng Lipid transfection mix 4.2 µl 4.58 µl M10 41.7 µl 45.83 µl
Table 2. Complex components. Per well and per well + 10% volumes of plasmid mix (Table 1), olfactory receptor plasmid (Rho-OR-pCI), and lipid transfection mix. M10 is added to quench the reaction following a 15 min incubation at room temperature.
- Pipette RTP1S-pCI, M3-R-pCI, pCRE-luc, and pSV40-RL plasmids into MEM medium per the volumes detailed in Table 1 to make the Plasmid mix (volumes indicated are per 96-well plate).
- Tap out the media on the cell plates.
- Pipette 50 µl of complex to each well and incubate overnight at 37 °C with 5% CO2.
4. Odor Stimulation
- Observe the transfected cells to ensure a proper confluency of 50-80% per well and return to incubator. NOTE: If cells are less than 50% confluent, firefly luciferase and renilla luciferase readings may be too low for measurement of receptor activation. Consider discarding the plate.
- Prepare 1 M stock solutions of each odor in DMSO.
- Prepare odor stimulation solutions in CD293 medium.
- For screening experiments, dilute stock solution of odor to 100 µM. Also prepare a no-odor control (CD293 only) in order to control for OR background activation. NOTE: For screening experiments, each OR/odor pair is tested only once per experiment. Because some odor diffusion across wells is possible, it is recommended to stimulate with one odorant for each plate.
- For dose response experiments, prepare seven 10-fold serial dilutions of odor stock solution in triplicate starting at 1 mM for each receptor. Also prepare the same odor dilutions in triplicate for empty vector-transfected cells in order to control for odor background activation. NOTE: For dose response experiments, each odor concentration treatment should be conducted in triplicate.
- Tap out the media on the cell plates.
- Pipette 25 µl of odor stimulation solution to each well and incubate for 4 hr.
5. Measuring OR Activity via Luciferase Assay
- Resuspend firefly luciferase substrate per the manufacturer’s instructions and store at -80 °C.
- Thaw 1 ml of firefly luciferase substrate per 96-well plate.
- Prepare fresh firefly luciferase reaction quencher and Renilla luciferase substrate reagent (5 µl luciferase quencher/Renilla luciferase substrate per 1 ml of buffer). NOTE: Approximately 1 ml of reagent is needed per 96-well plate.
- Prepare the luminescent microplate reader. Open the microplate reader software. Within the system icon:
- Under the "Pre-heating" Tab, check the box for "ON" and set the temperature of the machine to 25 °C.
- Under the "Dispenser" tab, prime each dispenser with 1,000 µl of 70% ethanol followed by 1,000 µl of distilled water. NOTE: Use separate aliquots of alcohol and water for each dispenser. Ethanol is used to disinfect the dispensers, and water removes the residual ethanol.
- Prime each dispenser with 1,500 µl of air (remove dispensers from liquid). NOTE: priming with air ensures that luciferase substrates are not diluted with residual water.
- Prime dispenser 1 with 1,080 µl of firefly luciferase substrate (from Step 5.2). Prime dispenser 2 with Renilla luciferase substrate (from Step 5.3). NOTE: Be careful not to cross-contaminate the luciferase substrates. Priming with luciferase substrates fills the dead space in the reagent dispensers.
- Set up the following protocol to read both firefly and Renilla luciferase luminescence. Within the software associated with the microplate reader, under the "File" menu, click on "New Task". Highlight "Protocols" and click on "Create New". In the next window, the circle next to "Standard Protocol" should be selected. Click "OK." Double click on "Procedure" on the left hand side of the screen.
- Dispense 10 µl of firefly luciferase substrate to all wells using dispenser 1. Under the "Actions" menu, click "Dispense". In the "Dispense Step" window, set: "Dispenser" to 1, "Priming" to none, "Dispense Volume" to 10 µl and "Rate" to 225 µl/sec. Click "OK".
- Shake the plate for 30 sec. Under the "Actions" menu, click "Shake". In the "Shake Step" window, set "Intensity" to Medium and "Duration" to 0:30 MM:SS. Click "OK".
- Read the luminescence of all wells for 0.5 sec per well. Under the "Actions" menu, click "Read". In the "Read Step" window, set: "Detection Method" to Luminescence, "Read Type" to Endpoint, "Integration Time" to 0:00:50 MM:SS:ss, "Filter Sets" to 1, "Emission" to Hole, "Optics Position" to Top, "Gain" to 135, and "Read Height" to 1.00 mm. Click "OK".
- Dispense 10 µl of Renilla luciferase substrate to all wells using dispenser 2. Set the conditions as in Step 5.6.1, except set "Dispenser" to 2.
- Shake plate for 30 sec. Set the conditions as in Step 5.6.2.
- Read luminescence of all wells for 0.5 sec per well. Set the conditions as in Step 5.6.3.
- Remove the lid from the 96-well plate and place the plate in the microplate reader. Start the program set up in Step 5.5 to read plate luminescence.
- Clean the reagent dispenser pumps. From the system icon under the "Dispenser" tab:
- Purge 1,000 µl of firefly luciferase substrate from the firefly luciferase dispenser into a recovery tube. NOTE: Firefly luciferase can be stored at -80 °C and reused.
- Prime each dispenser with 1,000 µl of distilled water, followed by 1,000 µl of 70% ethanol, and finally 1,500 µl of air (remove dispensers from liquid). NOTE: Water removes luciferase substrates from the reagent pumps, ethanol disinfects, and air dries any residual ethanol.
6. Data Analysis
- Data Export
- In the microplate reader software, double click on "Report/Export Builders" on the left side of the screen.
- Click on the button "New Export to Excel" and click "OK".
- Highlight Export1 and click "Edit".
- Under "Content" check "System Description", "Procedure", "Plate Description", and "Plate Layout Matrix". Include "Raw Data" and "Calculated Data".
- Under "Workflow", check "Autoexecute on completion of the procedure". Under Export Mode, check "All plates in the same workbook" and "As a new worksheet".
- Under "File" choose file name format and file location, and click "OK".
- Close the "Report/Export Builders" window.
- To obtain normalized luciferase values, divide the firefly luciferase luminescence reading for each well (Step 5.6.3) by the Renilla luminescence reading for each well (Step 5.6.6).
Representative Results
A primary screen tested 328 ORs against 26 odors at a concentration of 100 µM. This odor concentration has been demonstrated to effectively activate a large proportion of ORs with known ligands10. First, normalized luciferase activity was calculated by dividing the firefly luciferase reading by the Renilla luciferase reading. Next, baselined values were calculated by subtracting the normalized luciferase readings for the no odor control from the normalized luciferase readings for each well (Figure 1). Dose response curves were performed on 48 odorant/OR pairs randomly distributed across the range of baselined values, as indicated by colored bars in Figure 1. ORs were treated with 7 concentrations of odorant spanning 1 nM to 1 mM, and the resulting responses were fit to a sigmoidal curve using nonlinear regression. An odorant/OR was considered an agonist if it met three criteria: (1) the standard error of the logEC50 was less than 1 log unit; (2) the 95% confidence intervals for the top and bottom parameters of the curve did not overlap; and (3) the extra sums-of-squares test confirmed that the odorant activated the OR-containing cells significantly more than the control cells, which were transfected with an empty vector. Dose response results are summarized in Table 3.
These data were then used to determine how well assay measurements in a primary screen predict results from the dose response curve. Blue bars in Figure 1 correspond to pairs that were classified as agonists in a full dose response experiment, while red bars did not meet our three criteria outlined above. Values from the primary screen predicted results from the full dose response experiment (area under the receiver operating characteristic curve (AUC) = 0.68, p < 0.01, Mann-Whitney U test), indicating that our primary screen is a useful method to enrich for odorant/OR pairs that will be classified as agonists in a full dose response experiment (Figure 2).

Figure 1. Frequency of baselined luciferase values for a screen with a panel of olfactory receptors and odorants. Histogram of the frequency (Count) of baselined luciferase values calculated for each odorant/OR pair in the primary screen. As odorant/receptor activation pairs are sparse, the majority of the values are centered at zero and the large central distribution estimates the noise distribution for this assay. Colored bars indicate odorant/receptor pairs chosen for dose response analysis; blue bars are pairs that were classified as agonists based on the full dose response, and red bars are pairs that were not classified as agonists. Click here to view larger image.
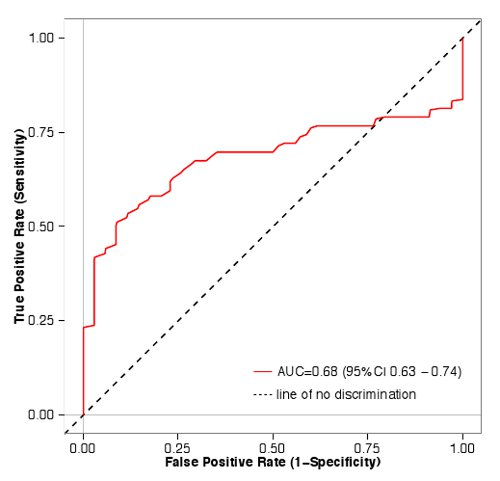
Figure 2. ROC curve for the odorant/receptor screen. 48 odorant/receptor pairs were classified as being agonists or as not being agonists. True positive rate (sensitivity) was then plotted against the false positive rate (1-specificity) using the R statistical package40. The area under the curve (AUC) is 0.68, indicating that odorant/receptor pairs with higher luciferase screen values are more likely to pass dose response than those with lower values. Click here to view larger image.
| Baselined Value | Dose Response |
| 0.051793067 | fail |
| 0.006376956 | fail |
| 0.331936398 | pass |
| 0.591006519 | pass |
| 0.049093369 | pass |
| 0.396788976 | pass |
| -0.013655743 | pass |
| 0.011080217 | pass |
| 0.004203349 | fail |
| 0.003975049 | fail |
| -0.077935718 | pass |
| -0.084488317 | pass |
| 0.030236078 | fail |
| -0.042963576 | fail |
| 0.031466406 | fail |
| 0.025897747 | fail |
| -0.030434651 | fail |
| -0.004122795 | fail |
| -0.010075533 | fail |
| 0.028883452 | fail |
| 0.019402373 | fail |
| 0.047508749 | fail |
| 0.00255344 | fail |
| 0.017221449 | fail |
| 0.340216655 | pass |
| -0.026912181 | fail |
| 0.037140428 | fail |
| 0.467763017 | pass |
| 0.097665337 | fail |
| 0.080657267 | pass |
| 0.172819211 | pass |
| 0.05568393 | pass |
| -0.106721064 | pass |
| 0.136614849 | pass |
| 0.457839849 | fail |
| 0.211751741 | fail |
| 0.1581464 | pass |
| -0.62099155 | pass |
| -0.066949491 | pass |
| -0.78712035 | pass |
| 0.752503007 | pass |
| 1.433407558 | pass |
| 0.475431098 | pass |
| 1.457936815 | pass |
| 0.048652537 | fail |
| 0.027196782 | fail |
| 0.129599842 | fail |
| -0.069781272 | fail |
| 0.016450039 | fail |
| -0.025639207 | fail |
| 0.158152141 | fail |
| -0.032570055 | fail |
| 0.140139926 | fail |
| -0.052030276 | fail |
| 0.657140133 | pass |
| 1.040410297 | pass |
| 0.164647156 | pass |
| 0.399588712 | pass |
| 0.188094387 | pass |
| 0.039371424 | pass |
| 0.016784352 | pass |
| 0.229959571 | pass |
| 0.238381997 | fail |
| 0.074118909 | fail |
| 0.423901128 | pass |
| 0.152621022 | pass |
| -0.109048046 | pass |
| 0.075301806 | pass |
| 0.395233972 | pass |
| 0.261892958 | pass |
| 0.156693306 | fail |
| 2.163418147 | pass |
| 3.649862104 | pass |
| 0.025716169 | pass |
| -0.033258008 | pass |
| -0.026984127 | fail |
| -0.338441868 | pass |
| 0.37398618 | pass |
Table 3. Olfactory receptor/odor pairs tested in dose response. Baselined luciferase values and dose response results (pass or fail) for 48 OR/odor pairs chosen from the screen. For 30 pairs tested in the screen twice, both baselined luciferase values are included.
Discussion
Odorant identity is encoded by olfactory receptor activation patterns, but receptor activation patterns, including which receptors are activated and to what degree, are known for fewer than 6% of human olfactory receptors1-11. Efforts to characterize olfactory receptors have been limited by their labor-intensive methods or applicability to only a subset of the olfactory receptor family17,23,24,33,34. The Hana3A heterologous expression system supports the robust expression of the majority of tested olfactory receptors, and can be used in conjunction with a cAMP-responsive luciferase reporter system to monitor olfactory receptor activation19,26,27. Performance of this assay in a 96-well format supports a number of high-throughput experimental designs, including screens to determine likely candidates for odorant/olfactory receptor pairs and dose-response curves to confirm interactions and assess how receptor activation levels are affected by intra- and inter-specific variations. Odorant/receptor pairs with higher activity values in a screen are more likely to demonstrate a significant dose response. These data suggest that this screening method is able to enrich for odorant/receptor pairs that will pass dose response, thereby facilitating the identification of odorant ligands and olfactory receptor activation patterns.
The success of this assay optimized for olfactory receptor analysis is dependent on several factors. All plasmid DNA must be prepared via an endotoxin-free protocol. Consistent olfactory receptor expression at the cell surface is critical. The Hana3A cell line stably expresses several accessory proteins that aid in OR expression, but co-transfection of RTP1S and M3-R enhances receptor expression and activation, respectively27. This combination of accessory protein expression results in the reliable expression of most olfactory receptors, allowing the comparison of OR activation among experiments and receptors. In addition, monitoring of cell confluency is important for obtaining consistent results. Assuming the cells in the original 10 cm2 dish are roughly 100% confluent, following the protocol described herein will result in reliable cell confluency throughout the experiment. Importantly, sufficient cells will be plated to obtain a measurable luciferase reading, but cells will not be over-grown, a condition which may affect receptor activation following odorant stimulation. Normalizing for constitutive renilla expression further controls not only for cell density, but also for transfection efficiency. A renilla luciferase reading more than 2.5 standard deviations below the mean may indicate cell loss. Cells should be plated uniformly to avoid dense plaques that detach more easily from the plate surface than sparser cells, and transfection and odorant solutions should be added gently to the side of the well to avoid detaching cells. Cell loss could also be due to cell death caused by odorant toxicity, a problem that may be circumvented by lowering odorant concentration, or excessive DMSO, which can be avoided by keeping DMSO concentrations below 0.5%. Finally, treating each receptor-expressing cell population with 1 µM forskolin, an adenylyl cyclase activator that causes luciferase reporter expression from the cAMP-responsive promoter, can serve as a positive control for the assay.
Although the assay described herein represents an improvement over alternative methods, including a high-throughput format and more general applicability to the mammalian olfactory receptor family, it has limitations. First, our in vitro assay lacks many components of an in vivo olfactory system, including odorant binding proteins, a mucosal layer, intracellular molecules and sniffing behaviors. Second, this method relies on a luciferase reporter system to measure olfactory receptor activation in contrast to common alternative methods that utilize calcium imaging. Recent work suggests that these two methods can produce conflicting results; indeed, a few olfactory receptors respond to a particular odorant when examined via calcium imaging but not luciferase assay41. Whether one assay type is more relevant to studies of human olfactory perception remains unclear, but both methods could be useful depending on context and receptor type. Third, while this functional expression system has successfully been used to express the majority of tested mammalian olfactory receptors, some ORs may not be amenable to expression using this system. If previously uncharacterized receptors fail to respond to an odor, it may be due to lack of expression at the cell surface rather than a lack of interaction between odorant and receptor. Receptor cell surface expression can be examined via immunofluorescence before drawing conclusions from negative assay results27,42. Finally, due to low background luciferase activity in no-odor conditions, our assay is not designed to detect inhibitory responses. To determine odor antagonists for olfactory receptors38,39, most receptors must first be stimulated with an odor in order to observe a reduction in luciferase activity.
Despite these limitations, this assay system has the ability to greatly increase data acquisition in the field of olfaction. First, the high-throughput 96-well format makes large-scale receptor and/or odor screens feasible. Second, its heterologous expression system is applicable to a variety of mammalian olfactory receptors. Third, luciferase activity can be used to measure olfactory receptor activation, which is valuable in describing the receptor activation patterns for a particular odorant. Fourth, previous results from similar in vitro assay systems predict human olfactory perception8,11,35. These characteristics are particularly important given the large size of the mammalian olfactory receptor family and our limited knowledge regarding the OR activation patterns elicited by specific odors. Broad application of this assay system optimized for olfactory receptor analysis will contribute to a more comprehensive picture of olfactory receptor/odorant interaction and the molecular basis of odor coding.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by R01 DC013339, R03 DC011373, and Ruth L. Kirschstein National Research Service Award T32 DC000014. A portion of the work was performed using the Monell Chemosensory Receptor Signaling Core, which is supported, in part, by funding from the NIH-NIDCD Core Grant P30 DC011735. The authors thank C. Sezille for help with data collection.
Materials
| Name of Material/ Equipment | Company | Catalog Number | Comments/Description |
| Hana3A cells | Avaiable from the Matsunami Laboratory upon request | ||
| RTP1S-pCI | Avaiable from the Matsunami Laboratory upon request | ||
| M3-R-pCI | Avaiable from the Matsunami Laboratory upon request | ||
| pCRE-luc | Agilent | 219076 | LUC |
| pSV40-RL | Promega | E2231 | RL |
| Minimum Essential Media, Eagle | Sigma Aldrich | M4655 | MEM |
| FBS | Life Technologies | 16000-044 | FBS |
| PBS (without Ca2+ and Mg2+) | Cellgro | 21-040-CV | PBS |
| Trypsin (0.05% Trypsin EDTA) | Life Technologies | 25300 | Trypsin |
| CD293 | Life Technologies | 11913-019 | CD293 |
| 96 well PDL white/clear plate | BD BioCoat | 356693 | plates |
| Lipid transfection reagent: Lipofectamine 2000 | Life Technologies | 11668-019 | Lipofectamine |
| Firefly luciferase substrate, firefly luciferase quencher/Renilla luciferase substrate: Dual-Glo Assay | Promega | E2980 | dual glo |
| Synergy S2 | BioTek | SLAD | BioTek S2 |
| Microplate reader software: Gen5 Data Analysis Software | BioTek | Gen5 | Gen5 |
| BIOSTACK | BioTek | BIOSTACK2WR | BioStack |
| Multiflo | BioTek | MFP | MultiFlo |
| 300ul GripTips | Integra | 4433 | GripTips |
| 12.5ul GripTips | Integra | 4414 | GripTips |
| 300ul GripTips ViaFlo96 | Integra | 6433 | XYZ tips |
| 12.5ul GripTips 384 XYZ | Integra | 6403 | XYZ tips |
| 384ViaFlo | Integra | 6030 | 384ViaFlo |
| TE buffer | Macherey Nagel | 740797.1 | |
| DMSO | Sigma Aldrich | D2650-100ML | DMSO |
| forskolin | Enzo Life Sciences | BML-CN100-0010 | FOR |
Riferimenti
- Wetzel, C. H., Oles, M., Wellerdieck, C., Kuczkowiak, M., Gisselmann, G., Hatt, H. Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes. The Journal of neuroscience the official journal of the Society for Neuroscience. 19 (17), 7426-7433 (1999).
- Spehr, M., et al. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 299 (5615), 2054-2058 (2003).
- Sanz, G., Schlegel, C., Pernollet, J. -. C., Briand, L. Comparison of odorant specificity of two human olfactory receptors from different phylogenetic classes and evidence for antagonism. Chemical senses. 30 (1), 69-80 (2005).
- Matarazzo, V., et al. Functional characterization of two human olfactory receptors expressed in the baculovirus Sf9 insect cell system. Chemical senses. 30 (3), 195-207 (2005).
- Jacquier, V., Pick, H., Vogel, H. Characterization of an extended receptive ligand repertoire of the human olfactory receptor OR17-40 comprising structurally related compounds. Journal of neurochemistry. 97 (2), 537-544 (2006).
- Neuhaus, E. M., Mashukova, A., Zhang, W., Barbour, J., Hatt, H. A specific heat shock protein enhances the expression of mammalian olfactory receptor proteins. Chemical senses. 31 (5), 445-452 (2006).
- Shirokova, E., et al. Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants. The Journal of biological chemistry. 280 (12), 11807-11815 (2005).
- Keller, A., Zhuang, H., Chi, Q., Vosshall, L. B., Matsunami, H. Genetic variation in a human odorant receptor alters odour perception. Nature. 449 (7161), 468-472 (2007).
- Menashe, I., et al. Genetic elucidation of human hyperosmia to isovaleric acid. PLoS biology. 5 (11), (2007).
- Saito, H., Chi, Q., Zhuang, H., Matsunami, H., Mainland, J. D. Odor coding by a Mammalian receptor repertoire. Science signaling. 2 (60), (2009).
- Jaeger, S. R., et al. A Mendelian Trait for Olfactory Sensitivity Affects Odor Experience and Food Selection. Current Biology. 23, 1-5 (2013).
- DeMaria, S., Ngai, J. The cell biology of smell. The Journal of cell biology. 191 (3), 443-452 (2010).
- Zhang, X., Firestein, S. The olfactory receptor gene superfamily of the mouse. Nature nauroscience. 5 (2), 124-1233 (2002).
- Glusman, G., Yanai, I., Rubin, I., Lancet, D. The complete human olfactory subgenome. Genome research. 11 (5), 685-702 (2001).
- Olender, T., Lancet, D., Nebert, D. W. Update on the olfactory receptor (OR) gene superfamily. Human Genomics. 3 (1), 87 (2008).
- Mombaerts, P. Genes and ligands for odorant, vomeronasal and taste receptors. Nature reviews. Neuroscience. 5 (4), 263-278 (2004).
- Malnic, B., Hirono, J., Sato, T., Buck, L. B. Combinatorial receptor codes for odors. Cell. 96 (5), 713-723 (1999).
- Araneda, R. C., Kini, a. D., Firestein, S. The molecular receptive range of an odorant receptor. Nature. 3 (12), 1248-1255 (2000).
- Saito, H., Kubota, M., Roberts, R. W., Chi, Q., Matsunami, H. RTP family members induce functional expression of mammalian odorant receptors. Cell. 119 (5), 679-691 (2004).
- Katada, S., Hirokawa, T., Oka, Y., Suwa, M., Touhara, K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. The Journal of neuroscience the official journal of the Society for Neuroscience. 25 (7), 1806-1815 (2005).
- Lu, M., Echeverri, F., Moyer, B. D. Endoplasmic Reticulum Retention, Degradation, and Aggregation of Olfactory G-Protein Coupled Receptors. Traffic. 4 (6), 416-433 (2003).
- McClintock, T. S., et al. Functional expression of olfactory-adrenergic receptor chimeras and intracellular retention of heterologously expressed olfactory receptors. Brain research. Molecular brain research. 48 (2), 270-278 (1997).
- Zhao, H. Functional Expression of a Mammalian Odorant Receptor. Science. 279 (5348), 237-242 (1998).
- Kajiya, K., Inaki, K., Tanaka, M., Haga, T., Kataoka, H., Touhara, K. Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants. The Journal of neuroscience the official journal of the Society for Neuroscience. 21 (16), 6018-6025 (2001).
- Krautwurst, D., Yau, K., Reed, R. R., Hughes, H. Identification of Ligands for Olfactory Receptors. Cell. 95, 917-926 (1998).
- Li, Y. R., Matsunami, H. Activation state of the M3 muscarinic acetylcholine receptor modulates mammalian odorant receptor signaling. Science signaling. 4 (155), (2011).
- Zhuang, H., Matsunami, H. Evaluating cell-surface expression and measuring activation of mammalian odorant receptors in heterologous cells. Nature. 3 (9), 1402-1413 (2008).
- Haddad, R., Khan, R., Takahashi, Y. K., Mori, K., Harel, D., Sobel, N. A metric for odorant comparison. Nature methods. 5 (5), 425-429 (2008).
- Veithen, A., Wilkin, F., Philippeau, M., Van Osselaer, C., Chatelain, P. Olfactory Receptors: From basic science to applications in flavors and fragrances. Perfumer and Flavorist. 35 (1), 38-40 (2010).
- Bozza, T., Feinstein, P., Zheng, C., Mombaerts, P. Odorant receptor expression defines functional units in the mouse olfactory system. The Journal of neuroscience the official journal of the Society for Neuroscience. 22 (8), 3033-3043 (2002).
- Oka, Y., Katada, S., Omura, M., Suwa, M., Yoshihara, Y., Touhara, K. Odorant receptor map in the mouse olfactory bulb: in vivo sensitivity and specificity of receptor-defined glomeruli. Neuron. 52 (5), 857-869 (2006).
- Zhao, H., Ivic, L., Otaki, J. M., Hashimoto, M., Mikoshiba, K., Firestein, S. Functional expression of a mammalian odorant receptor. Science. 279 (5348), 237-242 (1998).
- Touhara, K., et al. Functional identification and reconstitution of an odorant receptor in single olfactory neurons. Proceedings of the National Academy of Sciences of the United States of America. 96 (7), 4040-4045 (1999).
- Zhuang, H., Matsunami, H. Synergism of accessory factors in functional expression of mammalian odorant receptors. The Journal of biological chemistry. 282 (20), 15284-15293 (2007).
- McRae, J. F., Mainland, J. D., Jaeger, S. R., Adipietro, K. A., Matsunami, H., Newcomb, R. D. Genetic variation in the odorant receptor OR2J3 is associated with the ability to detect the "grassy" smelling odor, cis-3-hexen-1-ol. Chemical senses. 37 (7), 585-593 (2012).
- Adipietro, K. A., Mainland, J. D., Matsunami, H. Functional evolution of mammalian odorant receptors. PLoS genetics. 8 (7), (2012).
- Zhuang, H., Chien, M. -. S., Matsunami, H. Dynamic functional evolution of an odorant receptor for sex-steroid-derived odors in primates. Proceedings of the National Academy of Sciences of the United States of America. 106 (50), 21247-21251 (2009).
- Oka, Y., Nakamura, A., Watanabe, H., Touhara, K. An odorant derivative as an antagonist for an olfactory receptor. Chemical senses. 29 (9), 815-822 (2004).
- Oka, Y., Omura, M., Kataoka, H., Touhara, K. Olfactory receptor antagonism between odorants. The EMBO journal. 23 (1), 120-126 (2004).
- Fawcett, T. An introduction to ROC analysis. Pattern Recognition Letters. 27 (8), 861-874 (2006).
- Baghaei, K. A. Olfactory Receptors. Olfactory Recept. Methods Protoc. 1003, 229-238 (2013).
- Dey, S., Zhan, S., Matsunami, H. Assaying surface expression of chemosensory receptors in heterologous cells. Journal of visualized experiments JoVE. (48), (2011).

