Enhanced Reduced Representation Bisulfite Sequencing for Assessment of DNA Methylation at Base Pair Resolution
Summary
Enhanced Reduced Representation Bisulfite Sequencing is a method for the preparation of sequencing libraries for DNA methylation analysis based on restriction enzyme digestion combined with cytosine bisulfite conversion. This protocol requires 50 ng of starting material and yields base pair resolution data at GC-rich genomic regions.
Abstract
DNA methylation pattern mapping is heavily studied in normal and diseased tissues. A variety of methods have been established to interrogate the cytosine methylation patterns in cells. Reduced representation of whole genome bisulfite sequencing was developed to detect quantitative base pair resolution cytosine methylation patterns at GC-rich genomic loci. This is accomplished by combining the use of a restriction enzyme followed by bisulfite conversion. Enhanced Reduced Representation Bisulfite Sequencing (ERRBS) increases the biologically relevant genomic loci covered and has been used to profile cytosine methylation in DNA from human, mouse and other organisms. ERRBS initiates with restriction enzyme digestion of DNA to generate low molecular weight fragments for use in library preparation. These fragments are subjected to standard library construction for next generation sequencing. Bisulfite conversion of unmethylated cytosines prior to the final amplification step allows for quantitative base resolution of cytosine methylation levels in covered genomic loci. The protocol can be completed within four days. Despite low complexity in the first three bases sequenced, ERRBS libraries yield high quality data when using a designated sequencing control lane. Mapping and bioinformatics analysis is then performed and yields data that can be easily integrated with a variety of genome-wide platforms. ERRBS can utilize small input material quantities making it feasible to process human clinical samples and applicable in a range of research applications. The video produced demonstrates critical steps of the ERRBS protocol.
Introduction
DNA methylation at cytosine (5-methylcytosine) is an epigenetic mark critical in mammalian cells for a variety of biological processes, including but not limited to imprinting, X chromosome inactivation, development, and regulation of gene expression1-8. The study of DNA methylation patterns in malignant and other disorders has determined disease specific patterns and contributed to the understanding of disease pathogenesis and potential biomarker discoveries9-17. There are many protocols that interrogate the epigenome for DNA methylation status. These can be divided into affinity-based, restriction enzyme-based, and bisulfite conversion-based assays that utilize microarray or sequencing platforms downstream. Furthermore, there are a few protocols that bridge these general categories including, but not limited to, Combined Bisulfite Restriction Analysis18 and Reduced Representation Bisulfite Sequencing (RRBS19).
RRBS was originally described by Meissner et al.19,20. The protocol introduced a step to enrich GC-rich genomic regions followed by bisulfite sequencing, which resulted in quantitative base-pair resolution data that is cost effective21,22. The GC-rich regions are targeted by the MspI (C^CGG) restriction enzyme, and cytosine methylation is resolved by bisulfite conversion of cytosines (deamination of unmodified cytosines to uracil), followed by polymerase chain reaction (PCR) amplification. RRBS covered the majority of gene promoters and CpG islands in a fraction of the sequencing required for a whole genome; however RRBS had limited coverage of CpG shores and other intergenic regions of biological relevance. Several groups have published updated RRBS protocols since the original report that improve upon the methodology and resultant coverage of these genomic regions23-25. Enhanced Reduced Representation Bisulfite Sequencing (ERRBS) includes library preparation modifications and an alternate data alignment approach26 when compared to RRBS. ERRBS resulted in a higher number of CpGs represented in the data generated and increased coverage of all genomic regions interrogated26. This method has been used to resolve DNA methylation patterns in human patient and other animal specimens26-30.
The ERRBS protocol described offers details on all steps needed for completion and data was generated using representative human DNA (samples were obtained from previously reported, de-identified patient samples31, and a CD34+ bone marrow sample from a normal human donor). The protocol includes an automated size selection process, which reduces the processing time per sample and allows for increased accuracy in library size selection. The protocol combines a series of established molecular biology techniques. High molecular weight DNA is digested with a methylation-insensitive restriction enzyme (MspI) followed by end-repair, A-tailing, and ligation of methylated adapters. Size selection of the GC-rich fragments is followed by bisulfite conversion and PCR amplification prior to sequencing. Bisulfite conversion has been previously described32 and detailed review of data analysis and applications is beyond the scope of this paper, however recommendations and references are included for the readers’ use. The protocol can be performed over four days and is amenable to small input (50 ng or less) material amounts. The protocol as described yields data with high coverage per CpG site sufficient not only for differential methylation site and region determinations but also for epigenetic polymorphism detection as described by Landan, et al.33.
Protocol
NOTE: Institutional review board approval was obtained at Weill Cornell Medical College (protocol number 0805009783) and this study was performed in accordance with the Helsinki protocol.
NOTE: Please consult material safety data sheets for relevant materials before use (indicated throughout the protocol with “CAUTION”). Several of the reagents used are toxic and appropriate safety measures are advised (personal protective equipment and fume hood).
NOTE: All steps are performed at room temperature unless otherwise indicated throughout the protocol
1. Preparation and digestion of genomic DNA
- Prepare 50 ng of high quality genomic DNA (> 40 kilobases in size for human DNA) as starting material in 50 microliter (µl) of DNase-free water. Quantify the DNA using a fluorescence-based quantitation assay per manufacturer’s recommendations.
- Mix 50 ng of DNA with 2 µl of MspI (100,000 units/milliliter), 10 µl of appropriate 10X reaction buffer, and DNase-free water to bring the reaction to a total volume of 100 µl.
- Incubate the reaction at 37 °C in a thermal cycler or water bath for at least 18 hours.
- DNA extraction and precipitation
- Purify the digested DNA using a phenol-chloroform extraction.
- Add 200 μl of 10 millimolar (mM) tris(hydroxymethyl)aminomethane (Tris-Cl) pH 8.0 buffer to each reaction from step 1.3 to bring the volume to 300 μl.
- Add 150 µl of Tris- Ethylenediaminetetraacetic acid (TE) -saturated phenol and 150 µl of chloroform mix (1:1; CAUTION) to the DNA in a chemical hood and vortex briefly.
- Centrifuge at 20,800 x g in a microcentrifuge for 10 min at room temperature.
- Transfer top aqueous phase (approximately 300 µl) from last step into a new tube.
- Precipitate the digested DNA using ethanol precipitation.
- Add 1 µl of glycogen (20 milligrams/milliliter), 1/10 volume (30 µl) of 3 M pH 5.2 sodium acetate and 2.5 volumes (750 µl) of room temperature 100% ethanol. Mix by vortexing at high speed.
- Centrifuge at 4 °C for 45-60 min at 20,800 x g in a microcentrifuge.
- Remove ethanol quickly by inverting and dragging each tube over a paper towel with a quick flick of the wrist. Ensure that the DNA pellet remains on the side wall of the tube.
- Add 600 µl of room temperature 70% ethanol to each tube. Visualize the pellet as it dislodges from the tube side. Mix gently by inverting the tube five times.
- Centrifuge at 4 °C for 45-60 min at 20,800 x g in a microcentrifuge.
- Remove the ethanol by quickly inverting and dragging each tube over a paper towel with a quick flick of the wrist.
- Remove as much ethanol as possible by carefully removing any residual volume. Use a vacuum apparatus with a DNase-free 10 µl pipette tip at the end, and allow the pellet to air dry if necessary to fully eliminate any further residual ethanol.
NOTE: Avoid over drying the pellet since that will reduce the solubility upon resuspension in the next step. - Resuspend the DNA pellet into 30 µl of 10 mM Tris-Cl, pH 8.5 (Buffer EB).
NOTE: To avoid re-annealing of the “sticky” CG overhangs created by the MspI digestion, it is necessary to complete the protocol through the ligation step on the same day (protocol step 4).
- Purify the digested DNA using a phenol-chloroform extraction.
2. End-repair
- Transfer the 30 µl of MspI digested DNA (from step protocol 1.4.2.8) into a PCR tube on ice (4 °C) and add the reagents from Table 1.
- Incubate the end-repair reaction for 30 min at 20 °C in a thermal cycler with the heat lid on.
- Purify the DNA products to remove unincorporated dNTPs and other reaction reagents using a commercial column-based kit that binds of DNA in high-salt buffer and elutes DNA in low-salt buffer conditions per manufacturer’s recommendations. Elute the DNA products in 32 µl of Buffer EB.
3. A-tailing
- Transfer the 32 µl of DNA solution from step 2.3 into a PCR tube on ice (4 °C) and add the reagents listed in Table 2.
- Incubate the A-tailing reaction for 30 min at 37 °C in a thermal cycler with the heat lid on.
- Purify the DNA products to remove unincorporated dATPs and other reaction reagents using a commercial column-based kit that binds of DNA in high-salt buffer and elutes DNA in low-salt buffer conditions per manufacturer’s recommendations. Elute in 10 µl of Buffer EB.
4. Adapter ligation
- Transfer the 10 µl of A-tailed DNA into a PCR tube on ice (4 °C)
- Add the ligation reaction reagents to the DNA as detailed in Table 3 and adapters as detailed in Table 4.
- Incubate the ligation reaction overnight at 16 °C in a thermal cycler with the heat lid on.
- Purify the ligation products using a solid-phase reversible immobilization (SPRI) bead isolation protocol per manufacturer’s recommendations. For example, with Agencourt AMPure XP beads, use a 1.8X ratio of bead volume to sample volume (90 µl of beads for the 50 µl ligation reaction). Elute into 30 µl of DNase-free water.
- Ligated DNA can be stored at -20 °C before proceeding with size selection.
5. Size Selection
NOTE: Follow either section 5.1 for an automated size selection protocol or section 5.2 for a manual gel extraction size selection protocol. For samples with 25 ng or more of input DNA, an automated size selection protocol (using an instrument such as the Pippin Prep) can be used. Manual gel extraction is necessary for low DNA input amounts of 5-10 ng.
- Size selection using Pippin Prep for samples with 25 ng or higher of input DNA (Figure 2A) .
- Create a new protocol on the Pippin Prep for size selection.
- Select “2% DF Marker L” as the Cassette. Click the “Use internal standards” button. Verify that the “Ref lane” numbers match the lane numbers.
- Select “Range” as the collection mode for each lane. Enter 135 under "BP Start", 410 under "BP End", and 240 under "BP Pause" for each lane.
- Save the protocol.
- Add 10 µl of marker L to each 30 µl sample from the ligation reaction (step 4.3), bringing the total volume to 40 µl. Follow standard Pippin Prep protocol to prepare the gel cassette and instrument.
- Remove 40 µl of electrophoresis buffer from the sample wells, and load a 40 µl sample from step 5.1.5 into each of the 5 wells of the dye-free gel cassette.
NOTE: Dye-free, agarose gel cassettes are critical to size selecting adapter-ligated DNA fragments as the presence of ethidium bromide can alter the migration properties of the forked-adapter bound fragments. - Select the protocol created in step 5.1.1, and start the run.
- For each lane used collect 40 µl from the elution module when the run pauses at the 240 base pair (bp) point (lower library fraction: 135-240 bp). The instrument will pause individually for each lane. Repeat steps 5.1.5-5.1.7 for each lane as it pauses at 240 bp.
- Wash the elution module with 40 µl of fresh electrophoresis buffer by pipetting up and down three times. Discard the 40 µl of wash buffer. Repeat the wash step two additional times and remove all residual liquid from the elution module. Washing the elution module will decrease the amount of lower fraction DNA carried over into the higher fraction collection.
- Add 40 µl of fresh electrophoresis buffer and re-seal the elution module and resume the run.
- Collect 40 µl from each elution module into a new tube when the instrument indicates that the elution is complete. This elution contains the higher library fraction (240-410 bp). If running multiple gel cassettes, store size-selected samples at 4 °C before proceeding with bisulfite conversion.
- Size selection usingmanual gel extraction (Figure 2B)
- Prepare a 1.5% agarose gel with 0.2 µg/ml ethidium bromide (CAUTION).
- Load 50 bp and 100 bp ladders prepared with loading buffer in adjacent wells on both sides of the gel.
- Add 2 μl of 6X Orange G loading dye to the cleaned-up ligation products. Load the entire volume of each sample into individual wells, skipping at least one well between samples to avoid cross-contamination.
- Run the gel at 3.5 volts per centimeter until ladder is fully separated and the Orange G dye runs to the bottom of the gel (minimum of one hour)
- Slice off the ladders using a clean razor blade and visualize them on a UV transilluminator (CAUTION). Do not expose the samples to UV light. Mark the 150 bp, 250 bp, and 400 bp bands of the ladders using a razor blade or pipette tips. Marking the ladder bands will allow the excision of samples without exposing them to UV light.
- Align the ladder slices with the rest of the gel. Using the marked bands as reference, excise one slice containing the 150-250 bp library fraction (lower) and another slice containing the 250-400 bp library fraction (higher). Place each of the two slices into different tubes.
- Repeat the excision for all samples prepared using a clean razor blade for each gel lane used.
- Purify the lower and higher library fractions using a gel extraction kit following manufacturer’s protocols. Elute each sample extracted into 40 µl of EB. To ensure efficient PCR amplification of both the lower and higher library fractions in step 7, maintain the two fractions as independent samples for the bisulfite conversion step.
6. Bisulfite Conversion
- Set up a bisulfite conversion control per manufacturer’s recommendations (e.g., Universal Methylated Human DNA Standard kit). Treat the control in the same manner as the samples throughout the bisulfite conversion protocol.
- Perform bisulfite conversion using a commercial kit protocol per manufacturer’s recommendations. Use a commercial kit that has limited template degradation and DNA loss during treatment and cleanup, while resulting in high bisulfite conversion rates of the DNA. If using the EZ DNA Methylation Kit, perform the protocol per manufacturer’s recommendations with the following exception: incubate the samples in a thermal cycler using the following protocol: 55 cycles: 95 °C for 30 sec, 50 °C for 15 min. Hold at 4 °C.
- Elute the samples in 40 µl of DNase free water. Proceed with the enrichment PCR (step 7) on the same day that the bisulfite conversion is completed. Bisulfite-treated DNA is AU-rich and single-stranded, which reduces its stability.
- Confirm efficient bisulfite conversion of the control used via Sanger sequencing with non-methylated cytosines greater than 99% converted.
7. Enrichment PCR
- Prepare a PCR master mix using the reagents in Table 5 per library fraction (optimized for the use of FastStart Taq DNA Polymerase).
- Add the PCR master mix to each 40 µl bisulfite-converted library fraction. Mix by pipetting. Divide the 200 µl reaction into four PCR tubes with 50 µl each.
- Amplify the reactions in a thermal cycler with the following protocol: Set the heat lid to 100 oC. Initialize with a step of 94 °C for 5 min. Run 18 cycles of denaturing, annealing and extension/elongation steps: 94 °C for 20 seconds followed by 65 °C for 30 seconds followed by 72 °C for 1 min. Run a final extension/elongation step of 72 °C for 3 min and hold at 4 °C.
NOTE: 18 cycles of PCR is recommended for low input material quantities (less than 10 ng) and for first time users of the protocol. The protocol can be adjusted for a lower number of PCR cycles (as low as 14 cycles for 50 ng of input DNA; see Table 6).
8. Purify PCR reactions
- Combine each 4-reaction set (four 50 µl PCR reactions per library fraction).
- Purify the PCR products using a SPRI bead approach (or other approach which can remove unincorporated primers and other PCR reaction reagents) per manufacturer’s recommendations. The following steps have been optimized for Agencourt AMPure XP.
- Add 1.7X volume of SPRI beads (340 μl for 200 μl of PCR product) to the lower fraction amplification products.
- Add 1.1X volume of SPRI beads (220 μl for 200 μl of PCR product) to the higher fraction amplification products.
- Purify PCR products per manufacturer’s recommendations using 800 µl of 70% ethanol for wash steps.
- Elute into 50 µl of DNase-free water and add 1M Tris buffer to bring each library to 10 mM Tris-Cl, pH 8.5. Store libraries at -20 °C.
9. Library Quality Control
- Quantify libraries using a fluorescence-based quantitation assay selective for double-stranded DNA per manufacturer’s recommendations. Spectrophotometry-based measurements are not reliable. Expected concentrations measure 10 – 50 ng/µl for the lower library fraction and 3 – 15 ng/µl for the higher library fraction.
- Assess library sizes and quality using a bioanalyzer instrument and the High Sensitivity DNA Kit. Visualize library products and determine the average size of each library fraction. The lower library fraction typically has an average size between 180 and 210 bp. The higher fraction typically has an average size between 280 and 310 bp.
10. Prepare libraries for sequencing
- Calculate the molarity of each library fraction as follows:
- Size each library fraction using the trace obtained in step 9.2. For example, if using a bioanalyzer, use the “region” feature, and cover the beginning and end of the library fraction assessed.
- Record the average size of each library fraction in bp.
- Calculate molarity of each library fraction (Nanomolar concentration of DNA) using the following formula: Nanomolar concentration of DNA = [(ng/1000)/(bp*660)]*10^9 where ng is the concentration expressed in ng/μl (as measured in step 9.1) , bp is the average size of the library fraction, and 660 is the weight of a standard double stranded DNA base pair. For example: with a library that is 326 bp in size, at a concentration of 14.2 ng/μl, the Nanomolar concentration of DNA is 66 nM.
- Prepare 10 µl of a 2 nM solution of each library fraction. Dilute the library with DNase-free water.
- Pool the library fractions by combining 10 µl of the 2 nM lower fraction with 10 µl of the 2 nM higher fraction for each sample library prepared. This pool is the final ERRBS library for sequencing.
- Sequence the ERRBS library
- Load the libraries with a goal of optimal densities of 600,000 – 650,000 clusters per millimeter squared (suggested loading concentration of 7-8 picomolar).
- Sequence the libraries using a minimum of single-read 51 cycles sequencing on a HiSeq 2500 sequencer in high output mode.
- Use a designated control lane (see discussion for rationale).
11. Data Analysis
NOTE: Please refer to the Supplemental code files 1 and 2 for full details of commands and scripts recommended for use.
- Convert the base call files (.bcl files) to individual FASTQ files for each sample using the software provided by the sequencer’s manufacturer (example: CASAVA v1.8.2 for Illumina). If a sequencing core facility is used, this step may be provided and/or performed as an automated step on-board the sequencing computer.
- Filter the sequencing reads for reads that pass quality filtering (see Supplemental code file 1 for details). The CASAVA FASTQ file contains both reads that pass quality filtering and reads that do not pass quality filtering. Use a custom script that utilizes the <is filtered> element of the sequence identifier and keeps reads that have passed quality filtering.
- Trim adapter sequences from 3' end of the sequence reads in the filtered FASTQ files using a software tool that can remove adapter sequences, such as Flexbar 37. The minimum overlap length (-ao) set to 6, the minimum read length to keep after adapter removal (–m) set to 21, the cutoff for the number of allowed mismatches (-at) set to 2, and defaults for all other parameters. As an alternate to Flexbar, any software which can remove adapters can be used in step 11.3 (examples: cutadapt38, trimmomatic39).
- Use Bismark41 to align the filtered, adapter trimmed sequence reads to the bisulfite converted reference human genome hg19 (whole genome alignment approach) and determine the methylation context for each cytosine. Bismark is a customized short read mapping tool that aligns bisulfite treated reads to a bisulfite converted genome (where all ‘C’s are converted to ‘T’s) and returns methylation calls for cytosines in CpG, CHG, and CHH context. Typically set the seed length (-l) to the read length for alignment accuracy and use defaults for all other parameters.
- Sort aligned reads first by chromosome, then start position, and finally strand, after alignment is complete.
- Use custom scripts (see Supplemental code file 2 for script commands) to iterate over the sorted methylation calls output by Bismark to compute the percent methylation scores of bisulfite converted Cytosines (T’s; representing unmethylated C’s) and non-converted C’s (representing methylated C’s) for each cytosine methylation context (CpG, CHG, CHH), retaining only the cytosines that have at least phred quality score of 20 and have at least 10x coverage.
NOTE: The outputs are a methylation score file for each cytosine context with columns corresponding to: the position, strand, coverage, percent cytosine, and percent thymine, and for CHG and CHH context, a column for the next base (the H). - Use a custom script (see Supplemental code file 2 for script commands) to compute the conversion rates, from the methylation scores, and output the total of other C’s considered (CHG and CHH context), average conversion rate, and median conversion rate. Compute conversion rates for both strands independently as well as summarized over the entire library. Mean conversion rate is the fraction of C’s (converted and unconverted) in non-CpG context of the total number of C’s. Cytosines in CHG or CHH context are typically unmethylated and therefore present as thymine in the sequencing data.
- Generate a BAM file of the aligned reads using Bismark’s bismark2SAM_v5_xm.pl41 and SAMTOOLS42, and generate a wiggle format file using a custom script to convert the CpG methylation calls. The post-processed output can be converted to other formats, such as a bedgraph, and viewed in a genome browser such as the UCSC Genome Browser43 or IGV44.
Representative Results
Figure 1 provides an overview of ERRBS, highlighting key steps, which are explained throughout the protocol described. ERRBS libraries were prepared using 50 ng input DNA.
Evaluate the quality of the libraries prepared. Library production routinely yields fraction sizes of 150-250 bp and 250-400 bp (Figure 3A-C). Slight differences in library size distributions between samples are expected. Note that in both lower and higher library fractions there are very intense DNA sizes, indicative of enrichment of a particular sequence. MspI digestion results in the enrichment of a family of repetitive DNA sequences present in the human genome at 190 bp, 250 bp and 310 bp in the ERRBS libraries. These three repeats represent a characteristic signature of an ERRBS library20 (see Figures 3A-C and 3G). Representative libraries were sequenced on a next-generation sequencer using single-end reads. When loading at the recommended library concentration on an Illumina HiSeq 2500 sequencer, cluster densities of 500,000-700,000 per mm2 are expected. At this clustering density, 81.6% ± 3.14% (n = 81) of the clusters pass filter (Figure 4A). Due to the low complexity end of the library inserts (MspI recognition site: C^CGG), intensity values and quality scores recorded during sequencing are highly variable in the first three bases (Figure 4B-C), however, if an independent control lane is included (see discussion), 85% of bases will have quality scores of 30 or greater (Q30 values; Figure 4D).
Data alignment and cytosine methylation determination as described in the protocol yields base-pair resolution data (Table 7). For the human genome, a 51-cycle single-read sequencing run of an ERRBS library in one lane of a HiSeq 2500 in high output mode regularly generates 153,194,882 ± 12,918,302 total reads that after quality filtering and adapter trimming yields 152,231,183 ± 13,189,678 reads for input into the analysis pipeline. Average mapping efficiency for an ERRBS library is typically 62.95% ± 5.92% with representation of 3,183,594 ± 713,547 CpGs with a minimum coverage per CpG of 10x and an average coverage per CpG of 84.94 ± 16.29 (n = 100).
The ERRBS protocol is amenable to multiplexing (see Supplemental file 1: Protocol adaptation for multiplexed sequencing). Data from representative sequencing runs is summarized in Figure 5. Data from multiplexed sequencing runs (51-cycle single-read sequencing run; n = 128 for two libraries per lane; n = 11 for three libraries per lane; n = 11 for four libraries per lane) were compared to a full lane sequencing of an ERRBS library (51-cycle single-read sequencing runs; n = 100) as well as downsampling a single lane to simulate 50%, 33% and 25% of reads per lane (2, 3, and 4 sample multiplexing per lane respectively; n = 3). As the number of reads per sample decreases with the multiplexing factor, the number of CpGs covered at a minimum coverage of 10x and the coverage per CpG decreases as well (Figure 5 and Table 8). Mean conversion rates of non-CpG sites expected are 99.85% ± 0.04% (n = 400). Conversion rates lower than 99% may indicate less than optimal bisulfite conversion that can result in high rates of false methylation levels.
Data from an ERRBS library prepared from a representative human genomic DNA was analyzed in R 2.15.245 using the methylKit package26 (see Supplemental code file 1 for command details). The data can be visualized in commonly used genome browsers (Figure 6A). The cytosine methylation data is equally derived from both strands (Figure 6B) and ranges the entire spectrum of potential cytosine methylation levels (Figure 6C). Analysis of technical replicates from a representative human DNA sample yields high concordance between the data results (Figure 6D) and covers CpGs in a broad spectrum of genomic loci (Figure 6E and F and as previously described26). While technical replicas will yield high R2 values (greater than 97%), biological replicas will yield R2 values ranging from 0.92 to 0.9626, and comparing different human cell types will yield R2 values lower than 0.86 (data not shown).
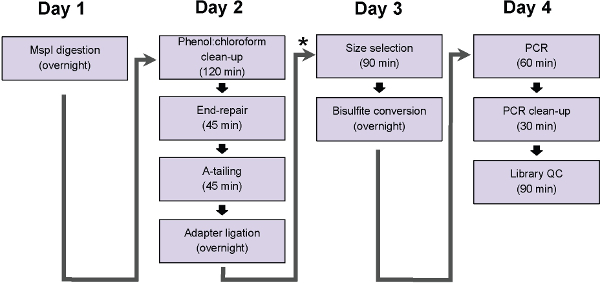
Figure 1: Flow chart of the ERRBS protocol steps. Chart represents steps, which can be completed in a traditional work day. * indicates a potential pause point (immediately following ligation clean up and before size selection, protocol step 5) at which samples can be frozen at -20 °C before proceeding with the duration of protocol.
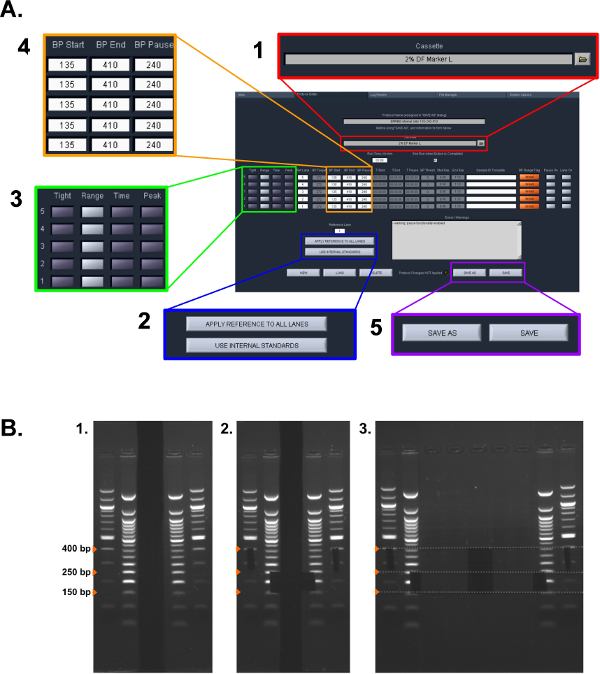
Figure 2: Size selection protocol. (A) Screen shot of settings used in the ERRBS Pippin Prep protocol (see protocol section 5.1.2 – 5.1.6): (1) Select Cassette type. (2) Select standard to be used. (3) Select the collection mode for each lane. (4) Enter the collection bp ranges. (5) Save the protocol. (B) Stages of the manual gel extraction used in protocol section 5.2: (1) Visualized gel ladders. (2) Marked Sizes for size selection using a razor blade. (3) Image of excised samples (lower fraction: 150-250 bp and higher fraction: 250-400 bp). Please click here to view a larger version of this figure.
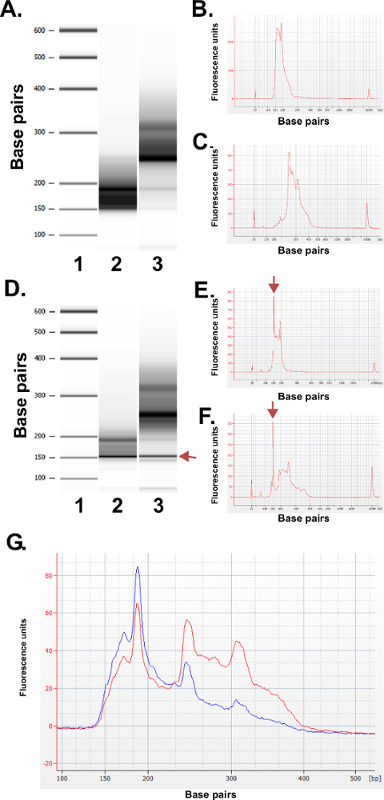
Figure 3: Quality control results for representative ERRBS libraries prepared from human DNA samples using a bioanalyzer machine. (A) Gel-like image showing a standard ladder (1), lower library fraction (135-240 bp fraction from Pippin Prep); 2) and the higher library fraction (240-410 bp fraction from Pippin Prep); 3). (B) Bioanalyzer electropherogram of the expected lower library fraction. (C) Bioanalyzer electropherogram of the expected higher library fraction. D–F) Representative data from a poor quality library prep. Gel-like image (D) of the standard ladder (1), lower library fraction (2) and the higher library fraction (3). The band at 150 bp marked with an arrow indicates excessive amounts of adapter. Electropherogram of the lower (E) and higher library fractions (F) with the excess adapter peaks at 150 bp (marked with arrows). (G) Bioanalyzer electropherogram of a pooled ERRBS library for sequencing. Red trace represents a high quality pooled library with equal representation of higher and lower fractions. Blue trace represents a pooled library not adequate for sequencing due to a lack of equal representation of the higher and lower fractions. Please click here to view a larger version of this figure.
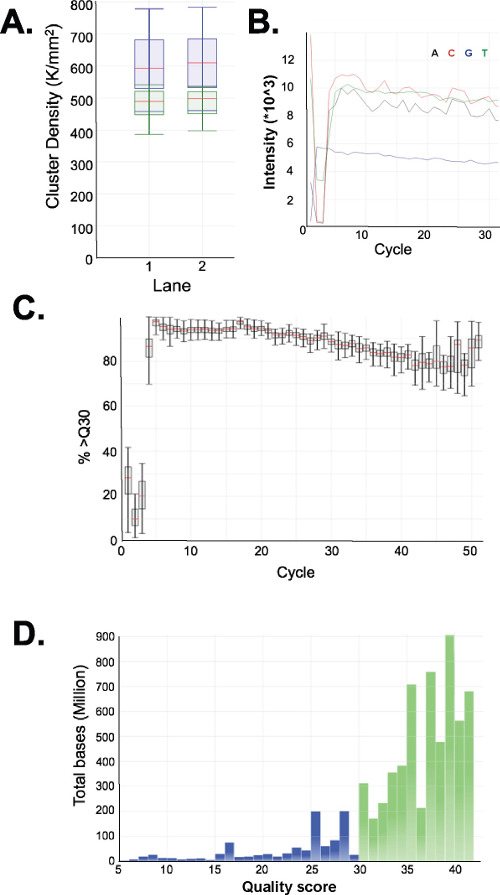
Figure 4: Sequencing charts for a representative ERRBS 51-cycle single-read sequencing run on a HiSeq 2500 sequencer in high output mode. (A) Cluster densities (K/mm2 = 1,000 clusters per millimeter squared; blue) and cluster densities passing filter (green) in two lanes with ERRBS libraries. (B) Typical intensities seen in the first 30 cycles in a lane with an ERRBS library. Note the CGG signature from MspI digestion in the intensities of the first three cycles. (C) Percentage of bases with a quality score of 30 or higher (%>Q30) for each cycle in one ERRBS lane. (D) Quality score distribution for all cycles in one ERRBS lane. Blue = less than Q30, Green = greater than or equal to Q30. In this lane, 84.7% of bases had quality scores of 30 or higher.
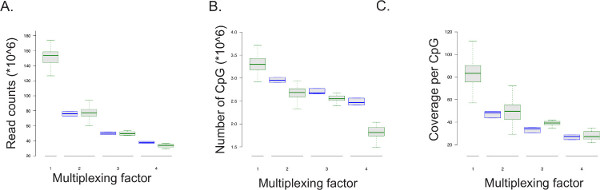
Figure 5: Sequencing output results. Box plots of experimental data from multiplexed and single sample per lane sequencing runs (displayed as green boxes) and of data derived by simulated downsampling from sequencing runs of three ERRBS libraries (displayed as blue boxes; sampled five times for each sequencing run) from 51-cycle single-read sequencing runs. The multiplexing factor corresponds to the number of ERRBS libraries sequenced per lane. 1 = whole lane or 100% of reads and represents data from a single ERRBS library per lane; 2 = 50% of lane and represents data from two ERRBS libraries per lane; 3 = 33% of a lane and represents data from three ERRBS libraries per lane; and, 4 = 25% of a lane and represents data from four ERRBS libraries per lane. (A) The read counts, or number of sequences analyzed, per multiplexing factor. (B) The number of CpG’s covered by the sequencing data per multiplexing factor. (C) The mean coverage per CpG per multiplexing factor. Please click here to view a larger version of this figure.
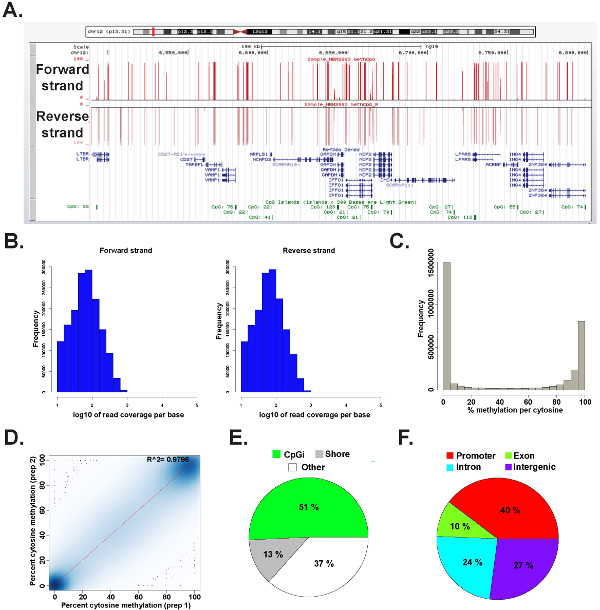
Figure 6: Representative data from an ERRBS library prepared from human genomic DNA. (A) University of California, Santa Cruz (UCSC) genome browser43 image of representative data from an ERRBS sequencing lane. The y-axis scale bar represents 0-100% methylation at each cytosine covered with a minimum of 10x. The top custom track represents the forward strand and the lower custom track represents the reverse strand. Shown is chr12:6,489,523-6,802,422 (hg19) inclusive of refseq genes and CpG islands within this genomic region. (B) Distribution histograms of CpG coverage along forward and reverse strands in a representative human CD34+ bone marrow sample. (C) Distribution histogram of CpG methylation levels along both strands in a representative human CD34+ bone marrow sample. (D) Correlation plot of CpG methylation levels from a representative technical replica of a human DNA sample. (E) Pie chart illustrating the proportions of CpGs covered in ERRBS which annotated to CpG islands (light green), CpG shores (gray) and other regions (white) in a representative sample prepared from human genomic DNA. (F) Pie chart illustrating the proportions of CpGs covered in ERRBS which annotated to gene promoters (red), exons (green), introns (blue) and intergenic regions (purple). Please click here to view a larger version of this figure.
| Reagent | Volume | Comment |
| 10x T4 DNA Ligase Reaction Buffer | 10 µl | |
| Deoxynucleotide triphosphate (dNTP) Solution Mix | 4 µl | mix of 10 mM of each nucleotide |
| T4 DNA Polymerase | 5 µl | 3,000 units/ml |
| DNA Polymerase I Large (Klenow) Fragment | 1 µl | 5,000 units/ml |
| T4 Polynucleotide Kinase | 5 µl | 10,000 units/ml |
| DNase-free water | 45 µl |
Table 1: End repair reaction reagents. Reagent names and quantities used in the end repair reaction (protocol step 2.1).
| Reagent | Volume | Comment |
| 10x reaction buffer | 5 µl | for example, NEBuffer 2 |
| 1 mM 2'-deoxyadenosine 5'-triphosphate (dATP) | 10 µl | |
| Klenow Fragment (3’→5’ exo-) | 3 µl | 5,000 units/ml |
Table 2: A-tailing reaction reagents. Reagent names and quantities used in the A-tailing reaction (protocol step 3.1).
| Reagent | Volume | Comment |
| 15 µM annealed adapters in DNase-free water | 3 µl | PE adapter 1.0 and PE adapter 2.0; see Table 4 for sequences and reference |
| 10x T4 DNA Ligase Reaction Buffer | 5 µl | |
| T4 DNA Ligase | 1 µl | 2,000,000 units/ml |
| DNase-free water | 31 µl |
Table 3: Adapter ligation reaction reagents. Reagent names and quantities used in the adapter ligation reaction (protocol step 4.2).

Table 4: Oligos used in the ERRBS protocol. List of oligos used throughout the ERRBS protocol in the ligation reaction (protocol step 4) and PCR amplification steps (protocol step 7). Please click here to view a larger version of this table.
| Reagent | Volume | Comment |
| 10x FastStart High Fidelity Reaction Buffer with 18 mM magnesium chloride | 20 µl | |
| 10 mM dNTP Solution Mix | 5 µl | |
| 25 µM PCR PE primer 1.0 | 4 µl | See Table 4 |
| 25 µM PCR PE primer 2.0 | 4 µl | See Table 4 |
| FastStart High Fidelity Enzyme | 2 µl | 5 units/µl FastStart Taq DNA Polymerase |
| DNase-free water | 125 µl |
Table 4: Oligos used in the ERRBS protocol. List of oligos used throughout the ERRBS protocol in the ligation reaction (protocol step 4) and PCR amplification steps (protocol step 7).
| Reagent | Volume | Comment |
| 10x FastStart High Fidelity Reaction Buffer with 18 mM magnesium chloride | 20 µl | |
| 10 mM dNTP Solution Mix | 5 µl | |
| 25 µM PCR PE primer 1.0 | 4 µl | See Table 4 |
| 25 µM PCR PE primer 2.0 | 4 µl | See Table 4 |
| FastStart High Fidelity Enzyme | 2 µl | 5 units/µl FastStart Taq DNA Polymerase |
| DNase-free water | 125 µl |
Table 5: PCR reaction reagents. Reagent names and quantities used in the PCR amplification reaction (protocol step 7.1).
| Protocol step | Reagent/protocol detail | Input DNA amount | ||
| 5-10 ng | 25 ng | 50 ng | ||
| 1 | MspI enzyme | 1 µl | 2 µl | 2 µl |
| MspI digest reaction volume | 50 | 100 | 100 | |
| 4 | Adapters in ligation reaction | 1 µl | 2 µl | 3 µl |
| Ligation reaction volume | 20 µl | 25 µl | 50 µl | |
| 5 | Size selection protocol | Manual gel only | Pippin Prep or manual gel | Pippin Prep or manual gel |
| 7 | PCR primer concentration | 25 µM | 25 µM | 10 µM for 14 cycles; 25 µM for 18 cycles |
| Number of PCR cycles | 18 | 18 | 14-18 | |
Table 6: Protocol step modifications for input material quantities ranging from 5-50 ng. Several steps throughout the protocol require modification of reagent quantities used to generate high quality libraries from various quantities of starting materials. Changes to key reagent quantities are included here. Adjust buffer and water volumes in reactions accordingly.
| Chr | Base | Strand | Coverage | freqC | freqT |
| chr1 | 10564 | R | 366 | 85.52 | 14.48 |
| chr1 | 10571 | F | 423 | 91.25 | 8.75 |
| chr1 | 10542 | F | 432 | 91.2 | 8.8 |
| chr1 | 10563 | F | 429 | 94.64 | 5.36 |
| chr1 | 10572 | R | 366 | 96.99 | 3.01 |
| chr1 | 10590 | R | 370 | 88.11 | 11.89 |
| chr1 | 10526 | R | 350 | 92 | 8 |
| chr1 | 10543 | R | 368 | 92.93 | 7.07 |
| chr1 | 10525 | F | 433 | 91.92 | 8.08 |
| chr1 | 10497 | F | 435 | 88.74 | 11.26 |
Table 7: Representative ERRBS data. After data alignment and cytosine methylation determination, base pair data is obtained. For each CpG covered, the alignment protocol as described will determine the genomic coordinate (columns: chr = chromosome, Base and Strand), the coverage rate of the specific locus (Coverage), and the rate of detection cytosine versus thymidine as percent (freqC and freqT respectively).
| Number of ERRBS libraries per lane | Mean number of uniquely aligned reads | Mean number of CpGs covered | Mean coverage per CpG |
| 1 | 152,231,184 ± 13,189,678 | 3,183,594 ± 713,547 | 85 ± 16 |
| 2 | 77,680,837 ± 7,657,058 | 2,674,823 ± 153,494 | 49 ± 9 |
| 3 | 49,938,156 ± 2,436,865 | 2,552,186 ±- 76,624 | 39 ± 2 |
| 4 | 34,457,208 ± 4,441,686 | 1,814,461 ± 144,339 | 28 ± 4 |
Table 8: Representative parameters from sequencing single and multiplexed ERRBS libraries. Shown is data per lane from 51-cycle single-read sequencing runs: mean and standard deviations of uniquely aligned reads, number of CpGs covered and coverage per CpG site obtained from sequencing single ERRBS libraries per lane (n = 100), two ERRBS libraries per lane (n = 128), three ERRBS libraries per lane (n = 11), and four ERRBS libraries per lane (n = 11).
Discussion
The protocol presented yields base-pair resolution data of cytosine methylation at biologically-relevant genomic regions. The protocol as written is optimized for 50 ng of starting material, however, it can be adapted to handle a range of input material (5 ng or more)26. This will require adjustments of some of the protocol steps as seen in Table 6. The ERRBS libraries are amenable to paired end sequencing and further genomic coverage can also be accomplished by sequencing reads longer than 51 cycles. Multiplexed sequencing will offer a lower cost protocol per sample, however, this will result in reduced coverage per CpG site represented in the data (Figure 5 and Table 8), and will not yield sufficient depth of coverage to perform analyses which require high coverage per CpG site (e.g. as described by Landan et al.33). Finally, this protocol (or any bisulfite-based protocol) cannot distinguish between methyl-cytosine and hydroxymethyl-cytosine46,47. However, the data generated can be integrated with other protocol results48,49 to delineate the different modifications, and other cytosine modifications recently reported50, should they be of interest.
High quality libraries will appear as shown in Figure 3A-C, and once pooled for sequencing yields a trace as shown in Figure 3G (red trace) representing equal molar contributions from both library fractions. Library preparation failure can result from any step during the procedure. If degraded DNA is processed it will result in libraries that are not enriched in MspI fragments and hence in low CpG coverage using the sequencing parameters described in this protocol. If an enzyme is non-functional or inadvertently excluded from one of the reactions, the protocol will not yield the expected library. If the ligation reaction is inefficient, adaptors are at a higher concentration than expected, and/or the primers concentration used is a limiting reagent for the final amplification steps, library failure can occur. Excess adapters (seen as a peaks at ~150 bp in bioanalyzer results; Figure 3D-F) in the library will also interfere with sequencing due to the indiscriminate clustering of both the library and excess adapters. While such a library may sequence apparently normally, a significant portion of the reads will be merely adapter sequences. If excess adapters are observed in a library, it is best to repeat the library preparation if material is available using optimal input material to adapter quantity ratios. Finally, to ensure efficient PCR amplification of the libraries, the lower and higher library fractions are maintained as separate samples throughout the bisulfite conversion and PCR enrichment steps. Failure to do so yields differential efficiency of amplification during the PCR reaction of higher and lower fractions (as seen in Figure 3G blue trace) and the potential for unequal representation of the respective genomic loci covered in each library fraction during sequencing. The user may opt to include a quantitative PCR step immediately after the bisulfite conversion for further titration of optimal PCR cycles needed to amplify the libraries being generated.
ERRBS library preparation protocol has several key steps in which specific reagents are recommended. At the end-repair step, the use of a four-nucleotide dNTP mix allows for end-repair of any products not containing the CG overhang, such as those resulting from MspI enzymatic star activity and sheared DNA fragments present in the original DNA sample. This results in improved CpG representation in the results. At the ligation step it is critical to use a high concentration ligase (2,000,000 units/ml) and methylated adapters to ensure that the ligation reaction is efficient and that the bisulfite conversion does not influence the adapter sequences essential for accurate data alignment. At the PCR step, using a polymerase capable of amplifying bisulfite-treated GC-rich DNA fragments is necessary for high specificity. Finally, to ensure elimination of excess adapters and primers, SPRI bead purification (for example: Agencourt AMPure XP) is recommended rather than column based assays for ligation and PCR product isolations.
In order to generate high quality data, it is important to ensure efficient bisulfite conversion. The control presented offers the user the ability to determine conversion efficiency prior to sequencing. As an alternative, a non-human DNA such as lambda DNA can be used as an internal control (spike-in). Due to the differences in species, this type of a control can be directly included in downstream sequencing (e.g. as used by Yu, et al.34). However, if the spike-in is utilized, it cannot be used to determine conversion efficiency prior to library sequencing unless uniquely amplified and independently sequenced prior to library sequencing. The conversion rates determined are based on the methylation status at non-CpG sites. This may not be appropriate for use in the context of high cytosine methylation in non-CpG context (for example embryonic stem cells) and parallel samples or other means of assessing for conversion efficiency can be utilized for this purpose.
There are a few caveats to address that are unique to the sequencing of ERRBS libraries. The first three bases of the library fractions sequenced are nearly uniformly non-random due to the MspI recognition cut site (C^CGG; see Figure 4B, C). This results in the potential for significant data loss due to low quality reads resulting from poor cluster localization in spite of apparent high cluster density during sequencing. To overcome this barrier, include a high complexity library in an independent lane (PhiX control or other library type) as a dedicated control lane. High complexity libraries have ends containing a balanced representation of A, C, T and G in the first four bases sequenced. Suitable control lanes include libraries such as RNA-seq, ChIP-seq, whole genome sequencing, or a control offered by the sequencing machine manufacturer (e.g. PhiX Control v3). When designated as a control lane for the respective sequencing run, it can serve as the basis for the matrix generation which is utilized during the first four bases of sequencing to detect cluster positions. The higher quality reads captured will raise the mean coverage per CpG site by 5.2 (n = 4). Alternatively, this technical difficulty can also be overcome using a dark sequencing approach as previously described23. Other sequencing criteria follow standard operating procedures per manufacturer’s protocols. Finally, the coverage per CpG chosen for data analysis will be guided by the user and in part by the biological questions of interest. 10x coverage threshold affords a high coverage analysis approach, however this threshold can be lowered should that be of interest.
A full discussion of ERRBS data analysis is beyond the scope of this article, however, differentially methylated cytosines and regions can be determined using open source tools 31,51-53. Additional analysis considerations and approaches have been well-described54,55, and the reader is encouraged to search the literature for tools most appropriate to the analysis planned.
Compared to other published methods, ERRBS offers a four-day protocol which when performed as described yields high rates of reproducibility. It has been validated compared to the gold standard MassARRAY EpiTYPER26, is cost-effective for high coverage data, and is adaptable for various input material amounts (favorable for clinical sample processing and other cell types of low frequency) and sequencing approaches. It offers base-pair resolution at biologically relevant loci and can be used in integrative analyses with other techniques profiling genome-wide transcription factor binding, chromatin remodeling, epigenetic marks and other cytosine modifications of interest. ERRBS data use in such studies can contribute to a comprehensive molecular approach and allow for high dimensional analyses in the study of biological models and human disease.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank all the authors of the original ERRBS report. We thank Mame Fall for technical assistance. We acknowledge the Weill Cornell Medical College Epigenomics Core for technical services and assistance. The work was supported by a Sass Foundation Judah Folkman Fellowship, an NCI K08CA169055 and ASH-AMFDP12005 to FGB, NIH R01HG006798 and R01NS076465, funding from the Irma T. Hirschl and Monique Weill-Caulier Charitable Trusts and STARR Consortium (I7-A765) to CEM, and an LLS SCORE grant (7006-13) to AMM.
Materials
| Name of Reagent/ Equipment | Company | Catalog Number | Comments/Description | URL |
| MspI | New England Biolabs | R0106M | 100,000 units/ml | https://www.neb.com/products/r0106-mspi |
| NEBuffer 2 | New England Biolabs | B7002S | Reaction buffer for MspI enzyme; protocol step 1.2 | https://www.neb.com/products/b7002-nebuffer-2 |
| Phenol solution | Sigma-Aldrich | P4557 | Equilibrated with 10 mM Tris HCl, pH 8.0; see safety and handling instructions at http://www.sigmaaldrich.com/catalog/product/sigma/p4557 | http://www.sigmaaldrich.com/catalog/product/sigma/p4557 |
| Chloroform | Sigma-Aldrich | C2432 | See safety and handling instructions at http://www.sigmaaldrich.com/catalog/product/sial/c2432 | http://www.sigmaaldrich.com/catalog/product/sial/c2432 |
| Glycogen | Sigma-Aldrich | G1767 | 19-22 mg/ml | http://www.sigmaaldrich.com/catalog/product/sigma/g1767 |
| NaOAc | Sigma-Aldrich | S7899 | 3M pH 5.2 | http://www.sigmaaldrich.com/catalog/product/sigma/s7899 |
| Ethanol | Sigma-Aldrich | E7023 | 200 proof, for molecular biology | http://www.sigmaaldrich.com/catalog/product/sial/e7023 |
| Buffer EB | Qiagen | 19086 | 10 mM Tris-Cl, pH 8.5 | http://www.qiagen.com/products/catalog/lab-essentials-and-accessories/buffer-eb |
| tris(hydroxymethyl)aminomethane (Tris) | Sigma-Aldrich | T1503 | prepare a 1M pH 8.5 solution | http://www.sigmaaldrich.com/catalog/product/sigma/t1503 |
| Tris- Ethylenediaminetetraacetic acid (TE) | Sigma-Aldrich | T9285 | Dilute to 1X buffer solution per manufacturer's recommendations | http://www.sigmaaldrich.com/catalog/product/sigma/t9285 |
| T4 DNA Ligase Reaction Buffer | New England Biolabs | B0202S | 10X concentration | https://www.neb.com/products/b0202-t4-dna-ligase-reaction-buffer |
| Deoxynucleotide triphosphate (dNTP) Solution Mix | New England Biolabs | N0447L | 10 mM each nucleotide | https://www.neb.com/products/n0447-deoxynucleotide-dntp-solution-mix |
| T4 DNA Polymerase | New England Biolabs | M0203L | 3,000 units/ml | https://www.neb.com/products/m0203-t4-dna-polymerase |
| DNA Polymerase I, Large (Klenow) Fragment | New England Biolabs | M0210L | 5,000 units/ml | https://www.neb.com/products/m0210-dna-polymerase-i-large-klenow-fragment |
| T4 Polynucleotide Kinase | New England Biolabs | M0201L | 10,000 units/ml | https://www.neb.com/products/m0201-t4-polynucleotide-kinase |
| QIAquick PCR Purification Kit | Qiagen | 28104 | Used for DNA product purification in protocol step 2.3 | http://www.qiagen.com/products/catalog/sample-technologies/dna-sample-technologies/dna-cleanup/qiaquick-pcr-purification-kit |
| 2'-deoxyadenosine 5'-triphosphate (dATP) | Promega | U1201 | 100 mM | http://www.promega.com/products/pcr/routine-pcr/deoxynucleotide-triphosphates-_dntps_/ |
| Klenow Fragment (3'→5' exo-) | New England Biolabs | M0212L | 5,000 units/ml | https://www.neb.com/products/m0212-klenow-fragment-3-5-exo |
| MinElute PCR Purification Kit | Qiagen | 28004 | Used for DNA product purification in protocol step 3.3 | http://www.qiagen.com/products/catalog/sample-technologies/dna-sample-technologies/dna-cleanup/minelute-pcr-purification-kit |
| T4 DNA Ligase | New England Biolabs | M0202M | 2,000,000 units/ml | https://www.neb.com/products/m0202-t4-dna-ligase |
| Methylation Adapter Oligo Kit | Illumina | ME-100-0010 | ||
| Agencourt AMPure XP | Beckman Coulter | A63881 | Used in protocol sections that implement magnetic bead purification steps (steps 4.3 and 8.2). Equilibrate to room temperature before use | https://www.beckmancoulter.com/wsrportal/wsrportal/wsr/research-and-discovery/products-and-services/nucleic-acid-sample-preparation/agencourt-ampure-xp-pcr-purification/index.htm?i=A63882#2/10//0/25/1/0/asc/2/A63882///0/1//0/ |
| Pippin Prep Gel Cassettes, 2% Agarose, dye-free | Sage Science | CDF2010 | with internal standards | http://store.sagescience.com/s.nl/it.A/id.1036/.f |
| Certified Low Range Ultra Agarose | Bio-Rad | 161-3106 | http://www.bio-rad.com/en-us/sku/161-3106-certified-low-range-ultra-agarose | |
| Tris-Borate-EDTA (TBE) buffer | Sigma-Aldrich | T4415 | http://www.sigmaaldrich.com/catalog/product/sigma/t4415 | |
| Ethidium bromide solution | Sigma-Aldrich | E1510 | 10 mg/ml | http://www.sigmaaldrich.com/catalog/product/sigma/e1510 |
| 50 bp DNA Ladder | NEB | N3236S | https://www.neb.com/products/n3236-50-bp-dna-ladder | |
| 100 bp DNA Ladder | NEB | N3231S | https://www.neb.com/products/n3231-100-bp-dna-ladder | |
| Gel Loading Dye, Orange (6X) | NEB | B7022S | https://www.neb.com/products/b7022-gel-loading-dye-orange-6x | |
| Scalpel Blade No. 11 | Fisher Scientific | 3120030 | http://www.fishersci.com/ecomm/servlet/fsproductdetail?position=content&tab=Items&productId=11876776 | |
| QIAquick Gel Extraction Kit | Qiagen | 28704 | http://www.qiagen.com/products/catalog/sample-technologies/dna-sample-technologies/dna-cleanup/qiaquick-gel-extraction-kit | |
| EZ DNA Methylation Kit | Zymo Research | D5001 | Used in protocol step 6.2 | http://www.zymoresearch.com/epigenetics/dna-methylation/bisulfite-conversion/ez-dna-methylation-kits/ez-dna-methylation-kit |
| EZ DNA Methylation-Lightning Kit | Zymo Research | D5030 | Alternative for step 6.2 | http://www.zymoresearch.com/epigenetics/dna-methylation/bisulfite-conversion/ez-dna-methylation-lightning-kit |
| Universal Methylated Human DNA Standard | Zymo Research | D5011 | Used as bisulfite conversion control | http://www.zymoresearch.com/epigenetics/dna-methylation/methylated-dna-standards/universal-methylated-human-dna-standard |
| FastStart High Fidelity PCR System | Roche | 03553426001 | http://lifescience.roche.com/shop/products/faststart-high-fidelity-pcr-system#tab-0 | |
| Qubit dsDNA High Sensitivity Assay Kit | Life Technologies | Q32854 | A fluorescence-based DNA quantitation assay; used in protocol steps 1.1, 9.1 and 10.1 | http://www.lifetechnologies.com/order/catalog/product/Q32854 |
| DynaMag-2 Magnet | Life Technologies | 12321D | https://www.lifetechnologies.com/order/catalog/product/12321D | |
| High Sensitivity DNA Kit | Agilent Technologies | 5067-4626 | http://www.genomics.agilent.com/en/Bioanalyzer-DNA-RNA-Kits/High-Sensitivity-DNA-Analysis-Kits/?cid=AG-PT-105&tabId=AG-PR-1069 | |
| 2100 Bioanalyzer | Agilent Technologies | http://www.genomics.agilent.com/en/Bioanalyzer-System/2100-Bioanalyzer-Instruments/?cid=AG-PT-106&tabId=AG-PR-1001 | ||
| PhiX Control v3 | Illumina | FC-110-3001 | http://www.illumina.com/products/phix_control_v3.ilmn | |
| HiSeq 2500 | Illumina | http://www.illumina.com/systems/hiseq_2500_1500.ilmn | ||
| Pippin Prep | Sage Science | http://www.sagescience.com/products/pippin-prep/ | ||
| Qubit 2.0 Fluorometer | Life Technologies | Q32872 | http://www.lifetechnologies.com/order/catalog/product/Q32872 | |
| TruSeq SR Cluster Kit v3-cBot-HS | Illumina | GD-401-3001 | http://www.illumina.com/products/truseq_sr_cluster_kit_v3-cbot-hs.ilmn | |
| TruSeq SBS Kit v3-HS | Illumina | FC-401-3002 | http://www.illumina.com/products/truseq_sbs_kit_v3-hs.ilmn | |
| TruSeq RNA Sample prep | Illumina | RS-122-2001 | Barcoded adapters used for multiplexing libraries; See Supplemental file for multiplexing protocol | http:/http://www.illumina.com/products/truseq-rna-access-kit.ilmn |
| Microcentrifuge | ||||
| Vortex Mixer | ||||
| Dry Block Heater | ||||
| Thermal Cycler | ||||
| Water Bath | ||||
| Gel electrophoresis system | ||||
| Electrophoresis power supply | ||||
| Gel doc | ||||
| UV or blue light transilluminator |
Riferimenti
- Jones, P. A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 13 (7), 484-492 (2012).
- Barlow, D. P. Genomic imprinting: a mammalian epigenetic discovery model. Annual Review Of Genetics. 45, 379-403 (2011).
- Thiagarajan, R. D., Morey, R., Laurent, L. C. The epigenome in pluripotency and differentiation. Epigenomics. 6 (1), 121-137 (2014).
- Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 447 (7143), 425-432 (2007).
- Hartnett, L., Egan, L. J. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis. 33 (4), 723-731 (2012).
- Smith, Z. D., Meissner, A. DNA methylation: roles in mammalian development. Nat Rev Genet. 14 (3), 204-220 (2013).
- Li, E., Bestor, T. H., Jaenisch, R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 69 (6), 915-926 (1992).
- Okano, M., Bell, D. W., Haber, D. A., Li, E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 99 (3), 247-257 (1999).
- Feinberg, A. P. Phenotypic plasticity and the epigenetics of human disease. Nature. 447 (7143), 433-440 (2007).
- Bock, C. Epigenetic biomarker development. Epigenomics. 1 (1), 99-110 (2009).
- Laird, P. W. The power and the promise of DNA methylation markers. Nat Rev Cancer. 3 (4), 253-266 (2003).
- How Kit, A., Nielsen, H. M., Tost, J. DNA methylation based biomarkers: practical considerations and applications. Biochimie. 94 (11), 2314-2337 (2012).
- Mikeska, T., Bock, C., Do, H., Dobrovic, A. DNA methylation biomarkers in cancer: progress towards clinical implementation. Expert Review Of Molecular Diagnostics. 12 (5), 473-487 (2012).
- Gyparaki, M. T., Basdra, E. K., Papavassiliou, A. G. DNA methylation biomarkers as diagnostic and prognostic tools in colorectal cancer. Journal of Molecular Medicine. 91 (11), 1249-1256 (2013).
- Figueroa, M. E., et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell. 17 (1), 13-27 (2010).
- Heyn, H., Mendez-Gonzalez, J., Esteller, M. Epigenetic profiling joins personalized cancer medicine. Expert review of Molecular Diagnostics. 13 (5), 473-479 (2013).
- Kulis, M., Esteller, M. DNA methylation and cancer. Advances in Genetics. 70, 27-56 (2010).
- Xiong, Z., Laird, P. W. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 25 (12), 2532-2534 (1997).
- Meissner, A., et al. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic Acids Res. 33 (18), 5868-5877 (2005).
- Gu, H., et al. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nat Protoc. 6 (4), 468-481 (2011).
- Bock, C., et al. Quantitative comparison of genome-wide DNA methylation mapping technologies. Nat Biotechnol. 28 (10), 1106-1114 (2010).
- Harris, R. A., et al. Comparison of sequencing-based methods to profile DNA methylation and identification of monoallelic epigenetic modifications. Nat Biotechnol. 28 (10), 1097-1105 (2010).
- Boyle, P., et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 13 (10), R92 (2012).
- Chatterjee, A., Rodger, E. J., Stockwell, P. A., Weeks, R. J., Morison, I. M. Technical considerations for reduced representation bisulfite sequencing with multiplexed libraries. Journal of Biomedicine & Biotechnology. 2012, 741542 (2012).
- Lee, Y. K., et al. Improved reduced representation bisulfite sequencing for epigenomic profiling of clinical samples. Biological Procedures Online. 16 (1), 1 (2014).
- Akalin, A., et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 8 (6), e1002781 (2012).
- Hatzi, K., et al. A Hybrid Mechanism of Action for BCL6 in B Cells Defined by Formation of Functionally Distinct Complexes at Enhancers and Promoters. Cell Reports. 4 (3), 578-588 (2013).
- Will, B., et al. Satb1 regulates the self-renewal of hematopoietic stem cells by promoting quiescence and repressing differentiation commitment. Nature Immunology. 14 (5), 437-445 (2013).
- Lu, C., et al. Induction of sarcomas by mutant IDH2. Genes Dev. 27 (18), 1986-1998 (2013).
- Kumar, R., et al. AID stabilizes stem-cell phenotype by removing epigenetic memory of pluripotency genes. Nature. 500 (7460), 89-92 (2013).
- Li, S., et al. An optimized algorithm for detecting and annotating regional differential methylation. BMC Bioinformatics. 14, S10 (2013).
- Patterson, K., Molloy, L., Qu, W., Clark, S. DNA methylation: bisulphite modification and analysis. Journal of Visualized Experiments. (56), 3170 (2011).
- Landan, G., et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 44 (11), 1207-1214 (2012).
- Yu, M., et al. Tet-assisted bisulfite sequencing of 5-hydroxymethylcytosine. Nat Protoc. 7 (12), 2159-2170 (2012).
- Goecks, J., Nekrutenko, A., Taylor, J., Galaxy, T. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 11 (8), R86 (2010).
- Dorff, K. C., et al. GobyWeb: simplified management and analysis of gene expression and DNA methylation sequencing data. PLoS One. 8 (7), e69666 (2013).
- Roehr, J. T., Dodt, M., Ahmed, R., Dieterich, C. Flexbar − flexible barcode and adapter processing for next-generation sequencing platforms. MDPI Biology. 1 (3), 895-905 (2012).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal, North America. 17 (1), 10-12 (2011).
- Bolger, A. M., Lohse, M., Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30 (15), 2114-2120 (2014).
- Needleman, S. B., Wunsch, C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 48 (3), 443-453 (1970).
- Krueger, F., Andrews, S. R. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 27 (11), 1571-1572 (2011).
- Li, H., et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25 (16), 2078-2079 (2009).
- Kent, W. J., et al. The human genome browser at UCSC. Genome Res. 12 (6), 996-1006 (2002).
- Thorvaldsdottir, H., Robinson, J. T., Mesirov, J. P. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in Bioinformatics. 14 (2), 178-192 (2013).
- Team, R. C. R. A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria. ISBN 3-900051-07-0, (2012).
- Nestor, C., Ruzov, A., Meehan, R., Dunican, D. Enzymatic approaches and bisulfite sequencing cannot distinguish between 5-methylcytosine and 5-hydroxymethylcytosine in DNA. BioTechniques. 48 (4), 317-319 (2010).
- Huang, Y., et al. The behaviour of 5-hydroxymethylcytosine in bisulfite sequencing. PLoS One. 5 (1), e8888 (2010).
- Yu, M., et al. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 149 (6), 1368-1380 (2012).
- Song, C. X., et al. Genome-wide profiling of 5-formylcytosine reveals its roles in epigenetic priming. Cell. 153 (3), 678-691 (2013).
- Ito, S., et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science. 333 (6047), 1300-1303 (2011).
- Akalin, A., et al. methylKit: a comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 13 (10), R87-1186 (2012).
- Stockwell, P. A., Chatterjee, A., Rodger, E. J., Morison, I. M. DMAP: Differential Methylation Analysis Package for RRBS and WGBS data. Bioinformatics. 30 (13), 1814-1822 (2014).
- Sun, D., et al. MOABS: model based analysis of bisulfite sequencing data. Genome Biol. 15 (2), R38 (2014).
- Bock, C. Analysing and interpreting DNA methylation data. Nat Rev Genet. 13 (10), 705-719 (2012).
- Rivera, C. M., Ren, B. Mapping human epigenomes. Cell. 155 (1), 39-55 (2013).

