Visualizing Monocarboxylates and Other Relevant Metabolites in the Ex Vivo Drosophila Larval Brain Using Genetically Encoded Sensors
Summary
Here we present a protocol to visualize the transport of monocarboxylates, glucose, and ATP in glial cells and neurons using genetically encoded Förster resonance energy transfer-based sensors in an ex-vivo Drosophila larval brain preparation.
Abstract
The high energy requirements of brains due to electrical activity are one of their most distinguishing features. These requirements are met by the production of ATP from glucose and its metabolites, such as the monocarboxylates lactate and pyruvate. It is still unclear how this process is regulated or who the key players are, particularly in Drosophila.
Using genetically encoded Förster resonance energy transfer-based sensors, we present a simple method for measuring the transport of monocarboxylates and glucose in glial cells and neurons in an ex-vivo Drosophila larval brain preparation. The protocol describes how to dissect and adhere a larval brain expressing one of the sensors to a glass coverslip.
We present the results of an entire experiment in which lactate transport was measured in larval brains by knocking down previously identified monocarboxylate transporters in glial cells. Furthermore, we demonstrate how to rapidly increase neuronal activity and track metabolite changes in the active brain. The described method, which provides all necessary information, can be used to analyze other Drosophila living tissues.
Introduction
The brain has high energy requirements due to the high cost of restoring ion gradients in neurons caused by neuronal electric signal generation and transmission, as well as synaptic transmission1,2. This high energy demand has long been thought to be met by the continuous oxidation of glucose to produce ATP3. Specific transporters at the blood-brain barrier transfer the glucose in the blood to the brain. Constant glycemic levels ensure that the brain receives a steady supply of glucose4. Interestingly, growing experimental evidence suggests that molecules derived from glucose metabolism, such as lactate and pyruvate, play an important role in the brain cells' energy production5,6. However, there is still some debate about how important these molecules are for energy production and which cells in the brain produce or use them7,8. The lack of appropriate molecular tools with the high temporal and spatial resolution required for this task is a significant issue that has prevented this controversy from being completely resolved.
The development and application of several engineered fluorescent metabolic sensors have resulted in a remarkable increase in our understanding of where and how metabolites are produced and used, as well as how the metabolic fluxes occur during basal and high neuronal activity9. Genetically encoded metabolic sensors based on Förster resonance energy transfer (FRET) microscopy, such as ATeam (ATP), FLII12Pglu700µδ6 (glucose), Laconic (lactate), and Pyronic (pyruvate), have contributed to our understanding of brain energy metabolism10,11,12,13. However, due to the high costs and sophisticated equipment required to conduct experiments on live animals or tissues, results in vertebrate models are still primarily limited to cell cultures (glial cells and neurons).
The emerging use of the Drosophila model to express these sensors has revealed that key metabolic features are conserved across species and their function can be easily addressed with this tool. More importantly, the Drosophila model has shed light on how glucose and lactate/pyruvate are transported and metabolized in the fly brain, the link between monocarboxylate consumption and memory formation, and the remarkable demonstration of how increases in neural activity and metabolic flux overlap14,15,16,17. The method presented here for measuring monocarboxylate, glucose, and ATP levels using genetically encoded FRET sensors expressed in the larval brain allows researchers to learn more about how the brain of Drosophila uses energy, which can be applied to the brains of other animals.
We show that this method is effective for detecting lactate and glucose in glial cells and neurons, and that a monocarboxylate transporter (Chaski) is involved in lactate import into glial cells. We also demonstrate a simple method for studying metabolite changes during increased neuronal activity, which can be easily induced by bath application of a GABAA receptor antagonist. Finally, we show that this methodology can be used to measure monocarboxylate and glucose transport in other metabolically significant tissues, such as fat bodies.
Protocol
1. Fly strain maintenance and larval synchronization
- To perform these experiments, use fly cultures raised at 25 °C on standard Drosophila food composed of 10% yeast, 8% glucose, 5% wheat flour, 1.1% agar, 0.6% propionic acid, and 1.5% methylparaben.
- To follow this protocol, use the following lines: w1118 (experimental control background), OK6-GAL4 (driver for motor neurons), repo-GAL4 (driver for all glial cells), CG-GAL4 (driver for fat bodies), UAS-Pyronic (pyruvate sensor), UAS-FLII12Pglu700µδ6 (glucose sensor), UAS-Laconic (lactate sensor), UAS-GCaMP6f (calcium sensor), UAS-AT1.03NL (ATP sensor) and UAS-Chk RNAi GD1829. All the lines expressing sensors or RNAi are in the w1118 genetic background.
- To obtain synchronized third instar wandering larvae, place 300 flies (3-5 days old, 100 males, 200 virgin females) of the desired genetic cross in the egg-laying chamber, which contains a 60 mm Petri dish covered with 1% agarose in phosphate-buffered saline solution (PBS). Place a drop of liquid/creamy yeast (1.5 cm diameter) in the center of the plaque to encourage flies to lay eggs (Figure 1).
- Keep the flies at 25 °C for 3 days replacing the agarose Petri dish with a fresh one and freshly dissolved yeast twice daily.
- Allow the flies to lay eggs for 4 h on the fourth day before changing the Petri dish. Remove this plaque. Then, for 3 h, allow the flies to lay eggs in a new agarose plaque with freshly dissolved yeast. Use these larvae in the experiments.
- After 3 h, remove the plaque containing the eggs and place it in a 25 °C incubator for 24 h.
- Collect 50 to 100 newly hatched larvae from this plaque (0 h after larval hatching) and place them in a plastic vial containing standard food. To allow the larvae to feed properly, ensure that the food is ground and soft. Use the larvae 96 h after the transfer.
2. Make the glass coverslips with poly-L-lysine
- Perform this step 1 h before the larval dissection. In a 6-well cell culture plate, place 25 mm glass coverslips (the diameter of the covers to be used depends on the recording chamber available in the microscope).
- Place a drop (300 µL) of poly-L-lysine in the center of each coverslip for 30 min at room temperature.
- Wash each coverslip 3x with distilled water, then 2x with a saline solution devoid of Ca2+ (the same solution used to dissect the larvae, see step 3.2). Install the covers in the recording chamber and fill it with Ca2+-free saline solution.
NOTE: Although the larval brain can adhere directly to the glass coverslips, the adhesion is weak, and the brain occasionally moves or displaces during the experiment due to the continuous flow of saline solution. The addition of the poly-L-lysine step reduces the risk of movements or even brain loss as a result of the washes.
3. Dissect the ventral nerve cord (VNC) and fat bodies (FBs)
- Gather the wandering third-instar larvae from the desired genetic cross (from step 1.7) and thoroughly wash them 3x with distilled water.
- Place the larvae in a glass dissection dish well containing 750 µL of nominally zero Ca2+ ice-cold saline solution composed of 128 mM NaCl, 2 mM KCl, 4 mM MgCl2, 5 mM trehalose, 5 mM HEPES, and 35 mM sucrose (pH = 6.7, measured with a pH meter and adjusted with 1 M NaOH and HCl 37% v/v).
NOTE: Setting the pH to 6.7 is critical because, as previously observed, monocarboxylate transport is highly dependent on the pH of the solution. In addition, 6.7 corresponds to the pH in the hemolymph of a third instar larva17,18. - Place the larva under a stereomicroscope and make a transverse cut across the back of the abdomen with a pair of forceps.
- Push the jaw with the forceps while turning the larvae inside out.
- Observe the ventral nerve cord (VNC) next to the jaw. Carefully remove the imaginal discs and remaining brain-caudal tissues.
- Separate the VNC with the central brain and optic lobes from the rest of the tissue by cutting the nerves. Transfer the VNC, picking it up with forceps from the remaining nerves, and placing it in the recording chamber containing the Ca2+-free recording solution (same solution from step 3.2). The VNCs will immediately adhere to the coverslips.
- To avoid interference from the remaining nerves, attach them at the bottom of the coverslip using forceps (the nerves will also be fluorescent depending on the driver line used) (Figure 2).
- To perform experiments in fat bodies (FBs) measuring glucose or monocarboxylate transport in another set of experiments, proceed to isolate the tissues following steps 3.1-3.4. Once the larvae are turned inside out, observe that the FB is a white, bilateral, flat tissue.
- Place the isolated FBs expressing the FRET sensors (obtained from a genetic cross using the appropriate drivers) in the recording chamber containing the recording solution without Ca2+.
NOTE: The FB is highly hydrophobic (it tends to float on the surface of the solution) and extremely vulnerable to manipulation with forceps, it must be handled with care. Any rough handling of this tissue will result in dead cells later (with a decreased fluorescence of the sensors). - Place the recording chamber containing VNCs/FBs on the microscope stage.
4. Live cell imaging
- To acquire images of the tissue expressing sensors, use an upright fluorescence microscope coupled to an emission splitting system and a CCD camera. Turn on the illumination system of the microscope 30 min before starting any experiment.
NOTE: Here we use a Spinning Disk fluorescence microscope equipped with a 20x/0.5 water immersion objective to observe both VNCs and FBs. Figure 2 also shows the use of a 40x/0.8 water immersion objective. - To visualize GCaMP6f fluorescence, set the excitation and emission wavelengths at 488 nm and 540 nm, respectively. For Laconic/Pyronic/ATP/glucose sensors, set the excitation wavelength at 440 nm and the emission wavelengths at 488 nm (mTFP, CFP) and 540 nm (Venus, YFP).
- Acquire images of 512 x 512 pixels size every 2 s for GCaMP6f and every 10-30 s for Laconic/Pyronic/ATP/glucose sensors. Determine the optimal exposure to the light source observing the quality of the image obtained. To follow this protocol, ensure that the images are of 300 ms exposure for Laconic and Pyronic sensors and 100-150 ms for the glucose and ATP sensors.
NOTE: The exposure can vary depending on the use of a confocal or normal fluorescence microscope. - Once the recording chamber is installed in the stage of the microscope, carefully place the water immersion objective and be sure that the objective remains submerged throughout the experiment. Keep the room temperature at 25 °C.
- Connect the recording chamber to a perfusion system and keep the tissues bathed in the recording solution containing 128 mM NaCl, 2 mM KCl, 1.5 mM CaCl2, 4 mM MgCl2, 5 mM trehalose, 5 mM HEPES, and 35 mM sucrose, pH 6.7 (measured with a pH meter and adjusted with NaOH and HCl).
- Maintain a constant flow of 3 mL/min of recording solution through the tissue using gravity, coupled to a low flux peristaltic pump to extract the liquid from the chamber while keeping the volume constant. To achieve the required flow, position the solution-containing tubes (50 mL plastic tubes) 25 cm above the microscope stage.
NOTE: This gravity-driven flow is used to prevent tissue movement caused by peristaltic pumps, allowing for more accurate image analysis later on. - Before any stimulation, maintain the recording solution flowing for 5-10 min to obtain a stable baseline of fluorescence from the sensors. Change the duration as necessary to obtain this baseline.
- Replace the flowing solution with the stimulation solution for 5 min to stimulate the VNC/FBs (with glucose, pyruvate, or lactate at the concentration described for each experiment dissolved in saline solution; see each figure).
NOTE: The duration of stimulation can be varied according to the needs of the experiment (for example, glucose pulses can be increased to 10 min to reach a plateau in neurons, as previously reported16). - To maintain osmolarity in the stimulation solution, rectify the sucrose concentration (a carbohydrate nonmetabolizable by the Drosophila brain) according to the concentration of the monocarboxylate or glucose added.
- Change the solutions using a simple system of valves. Be careful not to move the microscope stage.
- Expose the VNC to 80 µM picrotoxin (PTX, continuously flowing) to stimulate neuronal activity. This procedure increases the frequency of Ca2+ oscillations, making it possible to observe changes in metabolically relevant molecules (e.g., intracellular ATP).
- At the end of any experiment, thoroughly wash the complete flow system, including the tubes and the recording chamber, for at least 10 min with the saline solution and another 10 min with distilled water to eliminate any trace of the solutions used.
5. Image processing and data analysis
- To process the images obtained, proceed with the steps shown in Figure 3.
- Import the image (Laconic) to the ImageJ software. The image has two views of the sample: the left one is the mTFP signal and the one on the right is the Venus signal. Select a region that includes the cells to be analyzed and separate these images (mTFP and Venus) in a new window. Ensure that these images contain exactly the same area.
- Open the registration plugin and correct the small drift of the tissue that is normally observed in the experiment with the function rigid body. Perform this in both images (mTFP and Venus).
- Select at least 10 regions of interest (ROIs) in the cytosol of the corresponding 10 cells in the mTFP signal (488 nm) and transfer the selections to the Venus (540 nm) image.
- Select two or three ROIs close to the cells being analyzed but with no discernible signal (always within the VNC or tissue analyzed, see Figure 3). These are the background ROIs.
- With the ROIs selected in both images, obtain the mean grey value of each signal using the function measure. Transfer the data to a data sheet and subtract the average background value for each signal.
- Determine the mTFP fluorescence ratio over Venus (for Laconic and Pyronic). This value is computed differently than the other FRET sensors described here (where the ratio is usually YFP/CFP) because when bound to their respective substrates, both Laconic and Pyronic reduce their FRET efficiencies.
- Normalize the data dividing each recorded value by the baseline. In a separate data sheet, calculate the baseline by taking the mean value of the mTFP/Venus ratio during the 2 min preceding the stimulus.
- For the glucose and ATP measurements using FRET-based sensors, calculate the YFP/CFP ratio, and normalize the data as described in step 5.8.
Representative Results
For up to 1 h, this procedure allows for easy measurement of intracellular changes in the fluorescence of monocarboxylate and glucose sensors. As shown in Figure 4, Laconic sensors in both glial cells and motor neurons respond to 1 mM lactate at a similar rate at the start of the pulse, but motor neurons reach a higher increase over the baseline during the 5 min pulse, as previously demonstrated17. This lactate concentration was chosen because it is comparable to the levels found in the hemolymph of third-instar larvae. Similarly, when VNC-expressing glucose sensors were exposed to 5 mM glucose (a value similar to that measured in the hemolymph by us and others19), the sensor's signal increased at a similar rate in glial cells and neurons. During the glucose pulse, however, the signal in neurons increases more than in glial cells. This is consistent with previous observations of similar FRET sensor-expressing preparations16.
This preparation can be used to conduct experiments on undescribed monocarboxylate and glucose transporters expressed in Drosophila glial cells or neurons20. Here we exemplify the use of RNA-interference to show that Chaski (Chk), which was previously known to transport monocarboxylates in heterologous cells, transports lactate in glial cells (Figure 5). Under nutritional restriction conditions, this transporter was described as playing an important role in animal physiology and behavior21. We found that in glial cells, knocking out Chk reduced lactate transport compared to control VNCs exposed to 1 mM lactate (Figure 5). Other putative transporters can be tested to see if they can transport metabolites in physiological and pathological conditions.
To address the study of metabolic changes associated with neuronal activity, we developed a simple method to increase neuronal activity without using technically complex procedures such as optogenetics, which can interfere with FRET sensor measurements (Figure 6). The isolated VNC was exposed to Picrotoxin (PTX), a known GABAA receptor antagonist previously used by us and others17,22. The concentration used increases the frequency of calcium oscillations in neurons (as measured by changes in GCaMP6f fluorescence) as well as significant changes in metabolically relevant molecules like glucose, lactate, and pyruvate17. Moreover, PTX-induced increases in neuronal activity result in a transitory drop in ATP levels in the soma of motor neurons. This phenomenon has previously been described in vertebrate models in which neuronal activity was increased via electric stimulation or by using GABAA receptor antagonists23,24. As a result, this model can be used to track metabolic changes in specific subsets of neurons and glial cells, addressing the need for energy-related transporters that contribute to ATP production during high neuronal activity.
Glucose and monocarboxylate transport are also important in tissues or cells other than the brain. We show here the visualization of monocarboxylate (lactate/pyruvate) and glucose uptake in fat bodies, a metabolically relevant tissue present in the larvae and the adult fly that has several key functions such as lipid storage and metabolization25. The Laconic sensor is well expressed in FB, as shown in Figure 7, where the lipid droplets can be seen as tiny black spheres within the cell. This isolated tissue adheres well to the poly-L-lysine-coated coverslips, allowing for a long-term cell imaging procedure that is also easy to analyze. We observed a significant increase in the fluorescence of the glucose sensor when FB was exposed to increasing glucose concentrations (approximately 5 mM), which is close to the glucose concentration found in the hemolymph. Further exposure to higher concentrations of glucose (10 mM) does not result in the expected increase in sensor fluorescence. Finally, in FB, we were able to measure lactate and pyruvate import (Figure 7C,D). In this case, increasing the lactate and pyruvate concentrations causes a proportional increase in Laconic and Pyronic fluorescence (ranging from 0.1 to 1 mM). Higher monocarboxylate concentrations result in signal saturation inside the cells.
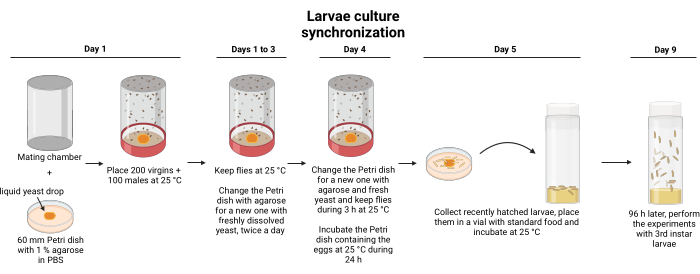
Figure 1: Synchronization of Drosophila larvae. The larval synchronization procedure is depicted graphically. Two hours before the process starts, 1% agarose plates must be prepared and changed twice daily until day 4. The hatching larvae must be collected with care. Please click here to view a larger version of this figure.
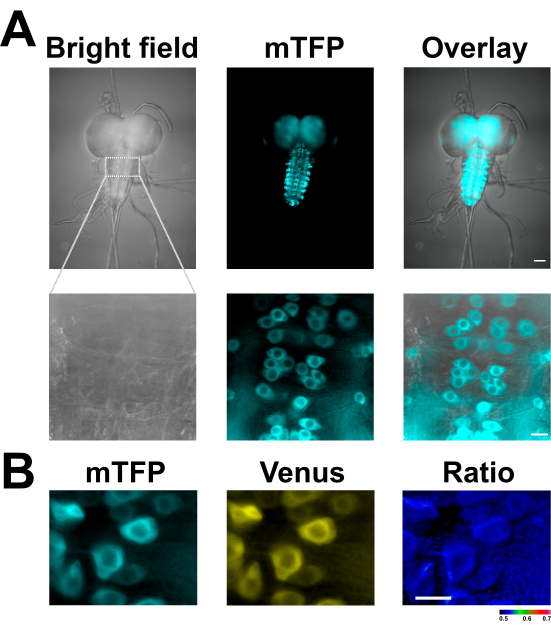
Figure 2: Isolated Drosophila larval brain expressing the lactate sensor Laconic. (A) A representative image of an isolated Drosophila third-instar larva's separated VNC adhered to a poly-L-lysine-coated coverslip and expressing the lactate sensor Laconic in motor neurons (OK6-GAL4). The images show a brightfield image of the VNC (left) and the sensor's mTFP fluorescence (middle panel) captured with a 20x water immersion objective. The images in the bottom panel are of the same VNC taken with a 40x water immersion objective. (B) Motor neurons from a VNC expressing the Laconic lactate sensor (OK6>Laconic) placed in a zero-lactate saline solution. The image depicts mTFP fluorescence (left), Venus fluorescence (center), and the ratio between the two fluorescence (right, produced using the Ratio Plus plugin of ImageJ software). Scale bars = 50 µm (A, top panel), 10 µm (A bottom panel, B). Abbreviations: VNC = ventral nerve cord; mTFP = monomeric teal fluorescent protein. Please click here to view a larger version of this figure.
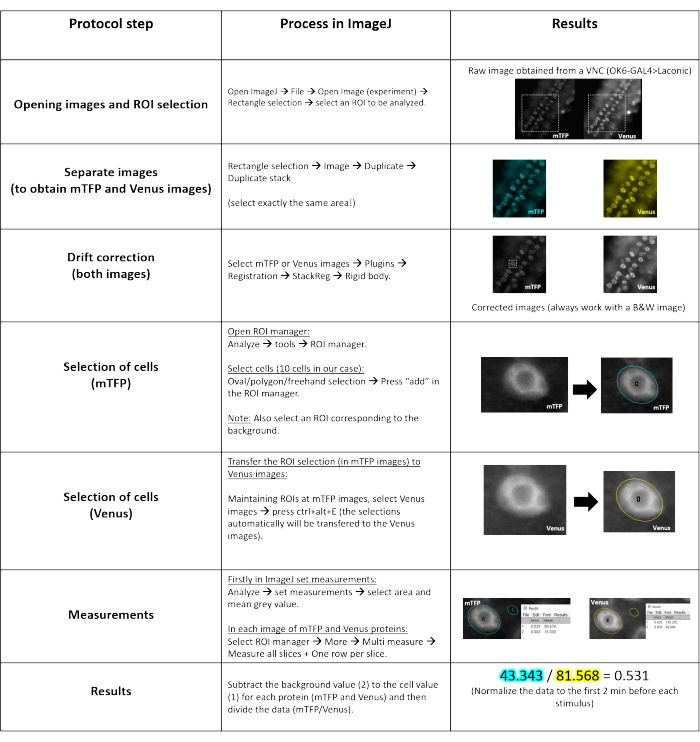
Figure 3: Step by step analyses of the images. A schematic illustration of the ImageJ software protocol for image analysis, displaying the process (first column), ImageJ commands (second column), and image processing outcomes (third column). The example starts with a raw image taken from a VNC expressing the Laconic lactate sensor in motor neurons (OK6-GAL4). Abbreviations: VNC = ventral nerve cord; mTFP = monomeric teal fluorescent protein. Please click here to view a larger version of this figure.
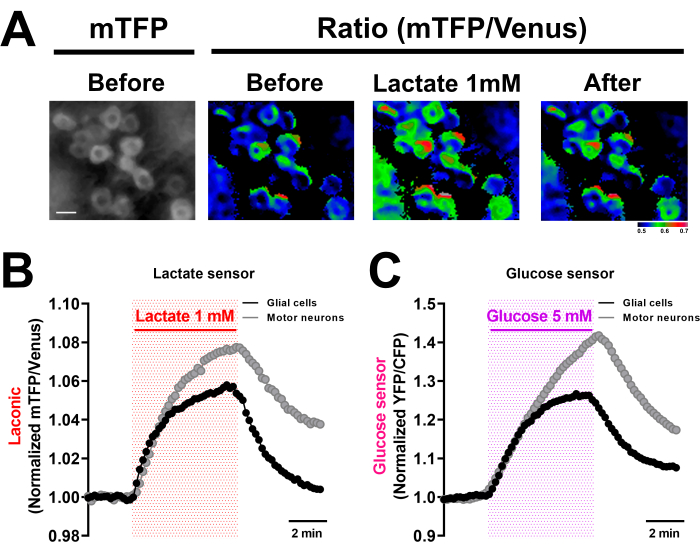
Figure 4: Monocarboxylate and glucose transport in neurons and glial cells of the Drosophila larval brain. (A) Representative images of VNC expressing the lactate sensor Laconic in motor neurons (OK6-Gal4>Laconic). Laconic fluorescence in motor neurons is seen before, during, and after a 5 min pulse of 1 mM of lactate. ImageJ's Ratio Plus plugin was used to create ratio images (mTFP/Venus). Scale bar = 10 µm. (B) Representative recordings of FRET signals from isolated VNCs expressing the Laconic lactate sensor in motor neurons (OK6-GAL4, grey lines) or glial cells (REPO-GAL4, black lines) exposed to 1 mM lactate for 5 min. The traces depict the adjusted FRET signals to the mean value collected 2 min before the lactate exposure. (C) Representative recordings of FRET signals from isolated VNCs expressing the glucose sensor FLII12Pglu700µδ6 in motor neurons (OK6-GAL4, grey lines) or glial cells (REPO-GAL4, black lines) exposed to 5 mM glucose for 5 min. The traces depict the adjusted FRET signals to the mean value collected two minutes before the glucose exposure. Abbreviations: VNC = ventral nerve cord; mTFP = monomeric teal fluorescent protein; FRET = Föster resonance energy transfer. Please click here to view a larger version of this figure.
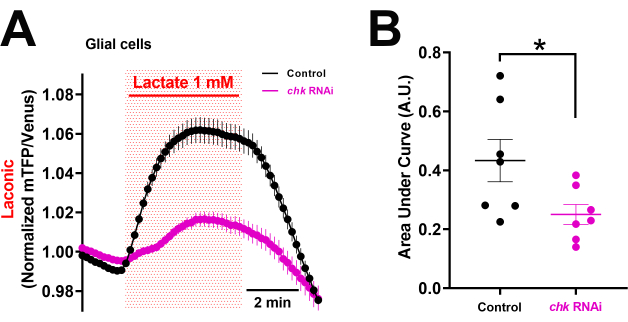
Figure 5: Lactate transport in glial cells of the Drosophila larval brain expressing Chaski RNAi. (A) Isolated VNC expressing the lactate sensor Laconic alone (genetic control, w1118, black line) or co-expressing an RNAi to knockdown Chaski (Chk) expression (chk-RNAi, magenta) were exposed to 1 mM Lactate for 5 min. For each condition, the traces represent the mean FRET value obtained from seven separate VNCs (70 cells per condition, normalized using the mean value obtained 2 min before being exposed to lactate). (B) The area under the curve in A is used to calculate intracellular lactate accumulation. For each condition, the values are the mean SE from 70 cells and seven independent experiments (control or chk-RNAi). The unpaired Student's t-test was used for statistical analysis (*p < 0.05). Abbreviations: VNC = ventral nerve cord; FRET = Föster resonance energy transfer. Please click here to view a larger version of this figure.
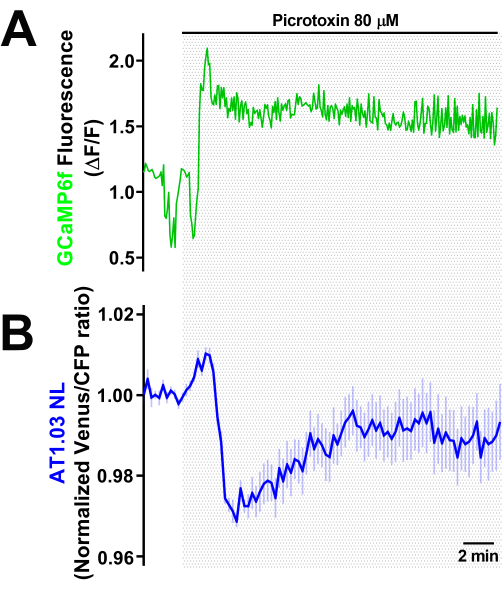
Figure 6: Changes in neuronal ATP levels induced by picrotoxin exposure. Recordings of the fluorescence captured from VNCs expressing the genetically encoded (A) calcium sensor GCaMP6f or (B, a different set of experiments) the ATP sensor AT1.03NL in motor neurons (OK6-GAL4) that were exposed to the GABAA receptor antagonist Picrotoxin (80 µM) during the time indicated by the line. The representative recording comes from a single motor neuron from a VNC (in A) or the mean ± SE from 10 cells from a single VNC (in B) normalized to the mean values obtained 2 min prior to PTX exposure. Abbreviations: VNC = ventral nerve cord; PTX = picrotoxin; SE = standard error; CFP = cyan fluorescent protein. Please click here to view a larger version of this figure.
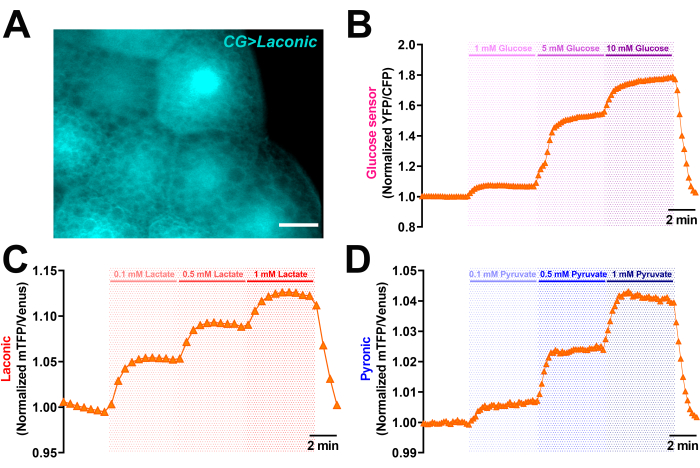
Figure 7: Transport of monocarboxylate and glucose in Drosophila fat bodies. (A) Image of the isolated fat body from third instar larvae expressing the lactate sensor Laconic (mTFP fluorescence). Scale bar = 10 µm. (B) Representative traces of the normalized signal captured in FB expressing the glucose sensor (FLII12Pglu700µδ6) exposed to glucose (1, 5, and 10 mM of glucose). Data were normalized to the first 2 min before glucose exposure. (C,D) Representative traces of the normalized signal obtained from FB expressing the lactate sensor (C) or the pyruvate sensor (D) exposed to 0.1-0.5-1 mM of lactate or pyruvate. The data were normalized to the first 2 min before lactate or pyruvate exposure. Abbreviations: FB = fat body; mTFP = monomeric teal fluorescent protein; YFP = yellow fluorescent protein; CFP = cyan fluorescent protein. Please click here to view a larger version of this figure.
Discussion
The use of the Drosophila model for the study of brain metabolism is relatively new26, and it has been shown to share more characteristics with mammalian metabolism than expected, which has primarily been studied in vitro in primary neuron cultures or brain slices. Drosophila excels at in vivo experiments thanks to the battery of genetic tools and genetically encoded sensors available that allows researchers to visualize in real time the metabolic changes caused by induced activity or even in response to a sensory stimulus.
The protocol described here shows how to measure monocarboxylate and glucose transport in Drosophila VNC glial cells and neurons in an ex-vivo preparation that allows the use of genetically encoded fluorescent metabolic sensors based on FRET microscopy to make time-lapse videos of metabolic changes in resting and active neurons. This protocol can be easily mounted and carried out in any lab using the Drosophila model without the use of complex machinery, using an epifluorescent microscope equipped with an emission-splitting system. It can also be scaled to more advanced equipment like a spinning disk, laser confocal, or two-photon microscopes (but a splitter to divide the light emission is required) to record specific cells more deeply located in the brain as occurs in adult Drosophila brain. As these types of metabolic signals are relatively slow, a fast or expensive video camera is not needed.
Additionally, the isolated VNC or FB preparation does not need a sophisticated method for movement correction sometimes needed for in vivo recordings. ImageJ plugins can satisfactorily correct most of the movements of the VNCs that are produced during the experiment. As a result, the increase in neuronal activity by the addition of PTX or the addition of relevant metabolites can be performed concurrently without the problems caused by natural body muscle contraction, as seen in other protocols (such as the fillet preparation)27.
We show representative experiments in which lactate and glucose transport is clearly observed at physiologically relevant concentrations of these metabolites (1 and 5 mM, respectively). From here, it is expected that any uncharacterized gene or previously reported protein can be tested for transport capacity in various conditions, such as using different models of neurodegenerative diseases or metabolic insults (high caloric diets or nutrient restriction). In this regard, we show that RNAi-mediated knockdown of a known monocarboxylate transporter reduces lactate transport in glial cells significantly. Interestingly, the signals obtained from Laconic and Pyronic sensors are highly sensitive to the changes in lactate or pyruvate in the media, allowing for an accurate measurement of the intracellular changes in these metabolites. This ample dynamic range of the signal seems to be different in mammalian cells where a nonlinear relationship between the signal of the sensor and the intracellular lactate concentration has been observed28.
This approach can provide answers to important aspects about where and when monocarboxylates and glucose are processed or utilized to generate energy in the brain during basal and high neuronal activity. The increase in neuronal activity caused by PTX can cause considerable alterations in energy-related metabolites, in a similar manner to what has been seen in cell cultures, brain slides, and living organisms23,24. Specifically, a surge in ATP production occurs minutes after an increase in neuronal activity, most likely due to glucose metabolization and the exchange of lactate and pyruvate between glial cells and neurons, as previously reported17. The study of the needs of specific cells for the supply of glucose and monocarboxylates, as well as the mechanisms underlying their transport or generation, can be made possible by manipulating the expression of certain genes in conjunction with the use of genetically encoded sensors.
Other metabolically important tissues can easily bind to poly-L-lysine-coated coverslips and metabolite changes be imaged for several minutes. Novel transporters mediating glucose or lactate transport in different tissues can be studied. Circulating glucose is transported to produce the disaccharide trehalose in larval fat bodies via mechanisms that are not fully understood, these methods could help to better understand this pathway. Furthermore, under nutritional restriction, fatty acids can be metabolized to produce ketone bodies in the FB and the transporters necessary to extrude them to the hemolymph are still unknown.
It is important to note that this protocol can be used to estimate the accumulation of a metabolite over time or to determine rates of increase in the fluorescence of a specific sensor between conditions, but not to formally consider changes in “concentration” because this would require an in-situ sensor calibration. The several cell layers that comprise the VNC in this case limit the diffusion of molecules, such as ionophores, necessary for this calibration. However, we advise following image processing guidelines that permit contrasting lactate levels between various organs or cells within an animal29,30.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
We thank all the members of the Sierralta Lab. This work was supported by FONDECYT-Iniciación 11200477 (to AGG) and FONDECYT Regular 1210586 (to JS). UAS-FLII12Pglu700µδ6 (glucose sensor) was kindly donated by Pierre-Yves Plaçais and Thomas Preat, CNRS-Paris.
Materials
| Agarose | Sigma | A9539 | |
| CaCl2 | Sigma | C3881 | |
| CCD Camera ORCA-R2 | Hamamatsu | – | |
| Cell-R Software | Olympus | – | |
| CG-GAL4 | Bloomington Drosophila Stock Center | 7011 | Fat body driver |
| Dumont # 5 Forceps | Fine Science Tools | 11252-30 | |
| DV2-emission splitting system | Photometrics | – | |
| Glass coverslips (25 mm diameter) | Marienfeld | 111650 | Germany |
| Glucose | Sigma | G8270 | |
| GraphPad Prism | GraphPad Software | Version 8,0,2 | |
| HEPES | Sigma | H3375 | |
| ImageJ software | National Institues of Health | Version 1,53t | |
| KCl | Sigma | P9541 | |
| LUMPlanFl 40x/0.8 water immersion objective | Olympus | – | |
| Methylparaben | Sigma | H5501 | |
| MgCl2 | Sigma | M1028 | |
| NaCl | Sigma | S7653 | |
| OK6-GAL4 | Bloomington Drosophila Stock Center | Motor neuron driver | |
| Picrotoxin | Sigma | P1675S | CAUTION-Fatal if swallowed |
| Poly-L-lysine | Sigma | P4707 | |
| Propionic Acid | Sigma | P1386 | |
| Repo-GAL4 | Bloomington Drosophila Stock Center | 7415 | Glial cell driver (all) |
| Sodium Lactate | Sigma | 71718 | |
| Sodium pyruvate | Sigma | P2256 | |
| Spinning Disk fluorescence Microscope BX61WI | Olympus | – | |
| Sucrose | Sigma | S0389 | |
| Trehalose | US Biological | T8270 | |
| UAS-AT1.03NL | Kyoto Drosophila Stock Center | 117012 | ATP sensor |
| UAS-Chk RNAi GD1829 | Vienna Drosophila Resource Center | v37139 | Chk RNAi line |
| UAS-FLII12Pglu700md6 | Bloomington Drosophila Stock Center | 93452 | Glucose sensor |
| UAS-GCaMP6f | Bloomington Drosophila Stock Center | 42747 | Calcium sensor |
| UAS-Laconic | Sierralta Lab | – | Lactate sensor |
| UAS-Pyronic | Pierre Yves Placais/Thomas Preat | – | CNRS-Paris |
| UMPlanFl 20x/0.5 water immersion objective | Olympus | – |
Riferimenti
- Vergara, R. C., et al. The energy homeostasis principle: neuronal energy regulation drives local network dynamics generating behavior. Frontiers in Computational Neuroscience. 13 (49), 1-18 (2019).
- Pulido, C., Ryan, T. A. Synaptic vesicle pools are a major hidden resting metabolic burden of nerve terminals. Science Advances. 7 (49), 1-9 (2021).
- Benton, D., Parker, P. Y., Donohoe, R. T. The supply of glucose to the brain and cognitive functioning. Journal of Biosocial Science. 28 (4), 463-479 (1996).
- Mergenthaler, P., Lindauer, U., Dienel, G. A., Meisel, A. Sugar for the brain: the role of glucose in physiological and pathological brain function. Trends in Neurosciences. 36 (10), 587-597 (2013).
- Boumezbeur, F., et al. The contribution of blood lactate to brain energy metabolism in humans measured by dynamic 13C nuclear magnetic resonance spectroscopy. The Journal of Neuroscience. 30 (42), 13983-13991 (2010).
- Baltan, S. Can lactate serve as an energy substrate for axons in good times and in bad, in sickness and in health. Metabolic Brain Disease. 30 (1), 25-30 (2015).
- Barros, L. F., Weber, B. CrossTalk proposal: an important astrocyte-to-neuron lactate shuttle couples neuronal activity to glucose utilization in the brain. The Journal of Physiology. 596 (3), 347-350 (2018).
- Yellen, G. Fueling thought: Management of glycolysis and oxidative phosphorylation in neuronal metabolism. The Journal of Cell Biology. 217 (7), 2235-2246 (2018).
- Koveal, D., Diaz-Garcia, C. M., Yellen, G. Fluorescent biosensors for neuronal metabolism and the challenges of quantitation. Current Opinion in Neurobiology. 63, 111-121 (2020).
- Imamura, H., et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proceedings of the National Academy of Sciences of the United States of America. 106 (37), 15651-15656 (2009).
- Takanaga, H., Chaudhuri, B., Frommer, W. B. GLUT1 and GLUT9 as major contributors to glucose influx in HepG2 cells identified by a high sensitivity intramolecular FRET glucose sensor. Biochimica et Biophysica Acta. 1778 (4), 1091-1099 (2008).
- San Martin, A., et al. A genetically encoded FRET lactate sensor and its use to detect the Warburg effect in single cancer cells. PloS One. 8 (2), 1-13 (2013).
- San Martin, A., et al. Imaging mitochondrial flux in single cells with a FRET sensor for pyruvate. PloS One. 9 (1), 1-9 (2014).
- Placais, P. Y., et al. Upregulated energy metabolism in the Drosophila mushroom body is the trigger for long-term memory. Nature Communications. 8, 1-14 (2017).
- Mann, K., Deny, S., Ganguli, S., Clandinin, T. R. Coupling of activity, metabolism and behaviour across the Drosophila brain. Nature. 593 (7858), 244-248 (2021).
- Volkenhoff, A., Hirrlinger, J., Kappel, J. M., Klambt, C., Schirmeier, S. Live imaging using a FRET glucose sensor reveals glucose delivery to all cell types in the Drosophila brain. Journal of Insect Physiology. 106 (1), 55-64 (2018).
- Gonzalez-Gutierrez, A., Ibacache, A., Esparza, A., Barros, L. F., Sierralta, J. Neuronal lactate levels depend on glia-derived lactate during high brain activity in Drosophila. Glia. 68 (6), 1213-1227 (2020).
- Geistlinger, K., Schmidt, J. D. R., Beitz, E. Human monocarboxylate transporters accept and relay protons via the bound substrate for selectivity and activity at physiological pH. PNAS Nexus. 2 (2), 1-8 (2023).
- Pasco, M. Y., Leopold, P. High sugar-induced insulin resistance in Drosophila relies on the lipocalin Neural Lazarillo. PloS One. 7 (5), 1-8 (2012).
- McMullen, E., Weiler, A., Becker, H. M., Schirmeier, S. Plasticity of carbohydrate transport at the blood-brain barrier. Frontiers in Behavioral Neuroscience. 14, 1-15 (2020).
- Delgado, M. G., et al. Chaski, a novel Drosophila lactate/pyruvate transporter required in glia cells for survival under nutritional stress. Scientific Reports. 8 (1), 1-13 (2018).
- Stilwell, G. E., Saraswati, S., Littleton, J. T., Chouinard, S. W. Development of a Drosophila seizure model for in vivo high-throughput drug screening. European Journal of Neuroscience. 24 (8), 2211-2222 (2006).
- Lerchundi, R., Huang, N., Rose, C. R. Quantitative imaging of changes in astrocytic and neuronal adenosine triphosphate using two different variants of Ateam. Frontiers in Cellular Neuroscience. 14 (80), 1-13 (2020).
- Baeza-Lehnert, F., et al. Non-canonical control of neuronal Energy Status by the Na(+) Pump. Cell Metabolism. 29 (3), 668-680 (2019).
- Mattila, J., Hietakangas, V. Regulation of carbohydrate energy metabolism in Drosophilamelanogaster. Genetica. 207 (4), 1231-1253 (2017).
- De Backer, J., Grunwald, I. A role for glia in cellular and systemic metabolism: insights from the fly. Current Opinion in Insect Science. 53 (100947), 1-8 (2022).
- Loganathan, S., Ball, H., Manzo, E., Zarnescu, D. Measuring glucose uptake in Drosophila models of TDP-43 proteinopathy. Journal of Visualized Experiments. (174), e62936 (2021).
- Dienel, G., Rothman, D. L. In vivo calibration of genetically encoded metabolite biosensors must account for metabolite metabolism during calibration and cellular volume. Journal of Neurochemistry. , (2023).
- Gandara, L., Durrieu, L., Behrensen, C., Wappner, P. A genetic toolkit for the analysis of metabolic changes in Drosophila provides new insights into metabolic responses to stress and malignant transformation. Scientific Reports. 9, 1-11 (2019).
- Gandara, L., Durrieu, L., Wappner, P. Metabolic FRET sensors in intact organs: Applying spectral unmixing to acquire reliable signals. BioRxiv. , (2023).

