High-Resolution Quantitative Immunogold Analysis of Membrane Receptors at Retinal Ribbon Synapses
Summary
The postembedding immunogold method is one of the most effective ways to provide high-resolution analyses of the subcellular localization of specific molecules. Here we describe a protocol to quantitatively analyze glutamate receptors at retinal ribbon synapses.
Abstract
Retinal ganglion cells (RGCs) receive excitatory glutamatergic input from bipolar cells. Synaptic excitation of RGCs is mediated postsynaptically by NMDA receptors (NMDARs) and AMPA receptors (AMPARs). Physiological data have indicated that glutamate receptors at RGCs are expressed not only in postsynaptic but also in perisynaptic or extrasynaptic membrane compartments. However, precise anatomical locations for glutamate receptors at RGC synapses have not been determined. Although a high-resolution quantitative analysis of glutamate receptors at central synapses is widely employed, this approach has had only limited success in the retina. We developed a postembedding immunogold method for analysis of membrane receptors, making it possible to estimate the number, density and variability of these receptors at retinal ribbon synapses. Here we describe the tools, reagents, and the practical steps that are needed for: 1) successful preparation of retinal fixation, 2) freeze-substitution, 3) postembedding immunogold electron microscope (EM) immunocytochemistry and, 4) quantitative visualization of glutamate receptors at ribbon synapses.
Introduction
Glutamate is the major excitatory neurotransmitter in the retina1. Retinal ganglion cells (RGCs), receiving glutamatergic synaptic input from bipolar cells2, are the output neurons of the retina that send visual information to the brain. Physiological studies showed that synaptic excitation of RGCs is mediated postsynaptically by NMDA receptors (NMDARs) and AMPA receptors (AMPARs) 3,4,5. Although excitatory postsynaptic currents (EPSCs) in RGCs are mediated by AMPARs and NMDARs3,5,6,7,8 , spontaneous miniature EPSCs (mEPSCs) on RGCs exhibit only an AMPARs-mediated component 4,5,9. However, reducing glutamate uptake revealed an NMDAR component in spontaneous EPSCs5, suggesting that NMDARs on RGC dendrites may be located outside of excitatory synapses . Membrane-associated guanylate kinases (MAGUKs) such as PSD-95 that cluster neurotransmitter receptors, including glutamate receptors and ion channels at synaptic sites, also exhibit distinct subsynaptic expression patterns 10,11,12,13,14.
Over recent decades, confocal immunohistochemistry and pre-embedding electron microscope (EM) immunohistochemistry have been employed to study membrane receptor expression. Although confocal immunostaining reveals broad patterns of receptor expression, its lower resolution makes it impossible to use to distinguish subcellular location. Pre-embedding EM studies in mammalian retina indicate that NMDAR subunits are present in postsynaptic elements at cone bipolar cell ribbon synapses 15,16,17. This is in apparent contrast to physiological evidence. However, diffusion of reaction product is a well-known artifact in the pre-embedding immunoperoxidase method. Hence, this approach does not usually give statistically reliable data and may exclude distinction between localization to synaptic membrane versus extrasynaptic membrane 18,19,20,21. On the other hand, physiological and anatomical data are consistent with a synaptic localization of AMPARs on RGCs 3,5,7,9,22. Thus, glutamate receptors and MAGUKs at retinal ribbon synapse are localized not only to the postsynaptic but also to the perisynaptic or extrasynaptic membrane compartments. However, a high-resolution quantitative analysis of these membrane proteins in a retinal ribbon synapse is still needed.
Here, we developed a postembedding EM immunogold technique to examine the subsynaptic localization of NMDAR subunits, AMPAR subunits and PSD-95 followed by estimating the number, density and variability of these proteins at synapses onto rat RGCs labeled using cholera toxin subunit B (CTB) retrograde tracing methods.
Protocol
Care and handling of animals were in accordance with NIH Animal Care and Use Committee Guidelines. Postnatal day (P) 15-21 Sprague-Dawley rats, injected with 1-1.2% CTB bilaterally through the superior colliculus, were maintained on a 12:12-hr light:dark cycle.
1. Retinal Tissue Fixation
- Assemble the following materials and tools: A dissecting microscope, 2 forceps with very fine tips, scissors, cellulose filter paper, plastic pipette and a microscope slide.
- Anesthetize the rat in a closed chamber with 2.0 ml halothane (an inhalant anesthetic). Determine adequate anesthetization by these methods: lack of withdrawal of rear paw after toe pinch, or lack of blink reflex. Then decapitate immediately with a guillotine. Remove the eyes with a pair of iris scissors and place in a glass dish containing 4% paraformaldehyde in 0.1 M phosphate buffer (PB) at pH 7.4.
- Using the dissecting microscope, remove the cornea by cutting off the front of the eyeball. Remove the lens and vitreous from the inner retinal surface with forceps.
- Peel the sclera with the two forceps until the retina is isolated from the eyecup.
- Cut the retina immediately into 100 – 200 µm-thick strips with a razor, and subject to pH-shift fixation.
- Fix retina strips in 4% paraformaldehyde in 0.1 M PB at pH 6.0 for 20 – 30 min and then in 4% paraformaldehyde plus 0.01% glutaraldehyde at pH 10.5 for 10 – 20 min at RT.
- After several washes in PB with 0.15 mM CaCl2 (pH 7.4 at 4°C), cryoprotect the retinal strips with glycerol (60 min each in 10%, 20%, 30%, then O/N in 30%) in 0.1 M PB prior to freeze substitution.
2. Freeze-substitution
NOTE: This freeze-substitution method is modified from an earlier published protocol 19,20. Also, it is crucial that the instruments are very cold (wear gloves); otherwise, the tissue may thaw partially when touched with the instruments. All of these steps are done within the AFS chamber and the instruments are never allowed to move above the rim of the chamber. Similarly, proper cooling of all chemicals used in the AFS is necessary.
- Plunge-freeze the retinal strips in liquid propane at -184°C in an EM cryopreservation instrument (CPC). Using a fine brush, place the samples on small pieces of double-stick tape attached to the metal stubs that go on the end of the plunging rods (wick off extra buffer using the brush).
- Plunge the rods into the liquid propane and keep there for about 5 sec, and then transfer to the automatic freeze-substitution instrument (AFS) using a small transport chamber that is filled with liquid nitrogen ('LN2 cooled cryo transfer container').
- After placing the frozen sample and instruments (forceps and scalpel) into the AFS, cool the instruments in the chamber for several minutes before touching the tissue, or cool them by placing the tips of the instruments for a few seconds into the small transport chamber (filled with liquid nitrogen ('LN2 cooled cryo transfer container')).
- Once in the AFS, remove the sample and tape from the stub using a fine scalpel. Also, keep the nitrogen gas flow control, TF (TF is described as a 'regulator control for LN2 vaporiser') open during these procedures.
- Prior to placing the frozen tissue into the flat-embedding holders (i.e., before setting up the AFS), cut a thin circle from a clear plastic sheet and place it into the bottom of the holder so as to line the bottom of each well. This allows relatively easier removal of the polymerized specimen blocks when finished.
NOTE: Previously, we used an alternative method to the flat embedding holders, and employing double Reichert+gelatin capsules (see Petralia and Wenthold, 1999) 20. - Use the following automatic sequence in the AFS (using instrument terminology): T1 = -90°C for 32 hr, S1 = increase temperature by 4°C/hr (11.3 hr), T2 = -45°C for 50 hr, S2 = increase temperature by 5°C/hr (9 hr), T3 = 0°C for 40 hr (total = 142.3 hr). It may take 2 – 4 hr to place samples in the AFS (at -90°C) prior to starting the timed sequence.
- Immerse the frozen sections in 1.5% uranyl acetate in methanol at -90°C for >32 hr in the AFS. Place two pieces of tissues (typically) in each of the seven wells in the flat-embedding aluminum holder (three fit into the AFS).
- Place the tissue+tape into the uranyl acetate/methanol in the wells and remove the tape later, just prior to beginning embedding medium (such as Lowicryl HM20) infiltration, if it is too difficult to remove the tissue from the tape.
- Then increase the temperature stepwise to -45°C (+4°C per hr; in the automatic sequence).
- Wash the samples three times in precooled methanol, by using a fine-tipped plastic pipette to remove the old solution from each flat-embedding holder, and then using another standard plastic pipette to add the precooled fresh methanol.
- Then, using the same method, infiltrate the samples progressively with low temperature embedding resins such as embedding medium (HM20/methanol at 1:1 and 2:1, each for 2 hr, followed by pure resin for 2 hr and then change the resins and keep O/N).
- Change the resin again the next day, adjusting the level of the resin to reach just to the top edge of the wells.
- Polymerize the samples (-45°C to 0°C in automatic sequence; +5°C per hr) with UV light for 40 hr.
- Then remove the samples from the AFS. Typically, sample blocks still show some pinkness in color. Place the samples close to the fluorescent light in the chemical fume hood, at RT O/N or longer until they appear completely clear.
3. Postembedding EM Immunogold Immunocytochemistry
NOTE: Postembedding immunocytochemistry is performed as described 23,24,25.
- Cut 1 µm sections with ultramicrotome, stain sections with 1% toluidine-blue, and examine them for section orientation; orient the tissue block to achieve the optimal transverse plane of sectioning.
- Cut 70 nm thick ultrathin sections with ultramicrotome and collect them on Formvar-carbon-coated nickel slot grids.
- Wash grids one time in distilled H2O followed by a Tris-buffered saline three times (TBS, 0.05 M Tris buffer, 0.7% NaCl, pH 7.6) wash.
- Incubate grids in 20 µl drops of 5% BSA in TBS for 30 min , and then in 20 µl drops of a mixture containing goat anti-CTB (1:3,000) and an antibody to one NMDAR subunit (anti-rabbit GluN2A 1:50, GluN2B 1:30), or an anti-rabbit GluA2/3 (1:30) in TBS-Triton (TBST, 0.01% Triton X-100, pH 7.6) with 2% BSA and 0.02 M NaN3 O/N at RT.
- Wash grids on three separate drops (20 µl) of TBS (pH 7.6) for 10, 10, and 20 min, followed by TBS (pH 8.2) for 5 min.
- Incubate grids for 2 hr on drops (20 µl) of a mixture containing donkey anti-rabbit IgG (1:20) coupled to 10 nm gold particles and donkey anti-goat IgG (1:20) coupled to 18 nm gold particles in TBST (pH 8.2) with 2% BSA and 0.02 M NaN3.
- Wash grids in 20 µl drops of TBS (pH 7.6) for 5, 5, and 10 min, then in ultrapure water and then dry them.
- Counterstain grids with 5% uranyl acetate and 0.3% lead citrate in distilled H2O for 8 and 5 min under dark conditions, respectively.
- For triple-labeling experiments, incubate grids O/N at RT with 20 µl drops of a mixture of anti-goat CTB (1:3,000), anti-mouse PSD-95 (1:100), and anti-rabbit GluN2A (1:50). Then incubate grids for 2 hr on 20 µl drops of a mixture of IgGs coupled to 18, 10, and 5 nm gold particles in TBST (pH 8.2) with 2% BSA and 0.02 M NaN3. Keep the same procedures as double labeling for washing and counterstaining between and after antibody incubation.
- Controls were performed in which the secondary antibodies were applied alone.
- View grids on an EM and digitalize images. Process final figures in Adobe Photoshop 6.0 only for brightness and contrast if it is necessary24.
4. Quantification
- Manually select areas of the inner plexiform layer (IPL) without holes or cracks at 8,000X magnification, then randomly photomontage the full depth of IPL at 25,000X magnification.
- Identify RGC dendrites at cone bipolar dyads when they a) contain the retrogradely transported CTB signal (large gold particles), b) exhibit well-defined membranes, clefts, and postsynaptic densities, c) contain at least two small gold particles within the PSD, or more than one small gold particle along the extrasynaptic plasma membrane.
- Collect 45 – 55 RGC dendrites, based on above criteria, for quantitative analysis.
- Measure the lengths of the PSD and the extrasynaptic plasma membrane of individual RGC dendrites with NIH ImageJ software. Count gold particles within 10 nm of the membrane as membrane-associated, based on an average thickness of 7 – 9 nm for the plasma membrane 26.
- Calculate particle density as the number of gold particles per linear micrometer (gold/µm). Measure the distance between the center of each gold particle and the middle of the PSD (for synaptic location) or the edge of the PSD (for extrasynaptic location).
- Perform statistical analysis by software. Use two-tailed t-tests to compare means. Conclude significance when P <0.05. Unless indicated otherwise, report values as mean ± SEM18.
Representative Results
The results presented here demonstrate strikingly different subsynaptic localization patterns of GluA 2/3 and NMDARs on RGC dendrites in rat retina, as described previously 24,25. 77% of GluA 2/3 immunogold particles in RGC dendritic profiles were located within the PSD (Figure 1A), similar to most central synapses. However, NMDARs were located either synaptically or extrasynaptically. 83% of GluN2A immunogold particles were localized in the PSD (Figure 1C, 2A, 2B), preferentially at OFF synapses, and the gold particles were more concentrated at the center of the PSD (Figure 2C). In contrast, the large majority of particles labeling for GluN2B (96%) were located outside the PSD (Figure 1B, 2A, 2B), distributed primarily along the extrasynaptic plasma membrane, with the peak particle density at 180 nm from the edge of the PSD (Figure 2B). In particular, GluN2B exhibited a preference for ON synapses. In a subset of experiments, simultaneous labeling of GluN2A, PSD-95 and CTB indicated that GluN2A and PSD-95 were colocalized in the PSD of OFF RGC dendrites (Figure 1D), where NR2A was in the dendritic membrane while PSD-95 was beneath the membrane, suggesting that they are anchored at the PSD. A significant difference in gold density between perisynaptic membrane and mitochondrial membranes was observed, suggesting a specific labeling in the former (Figure 2D).
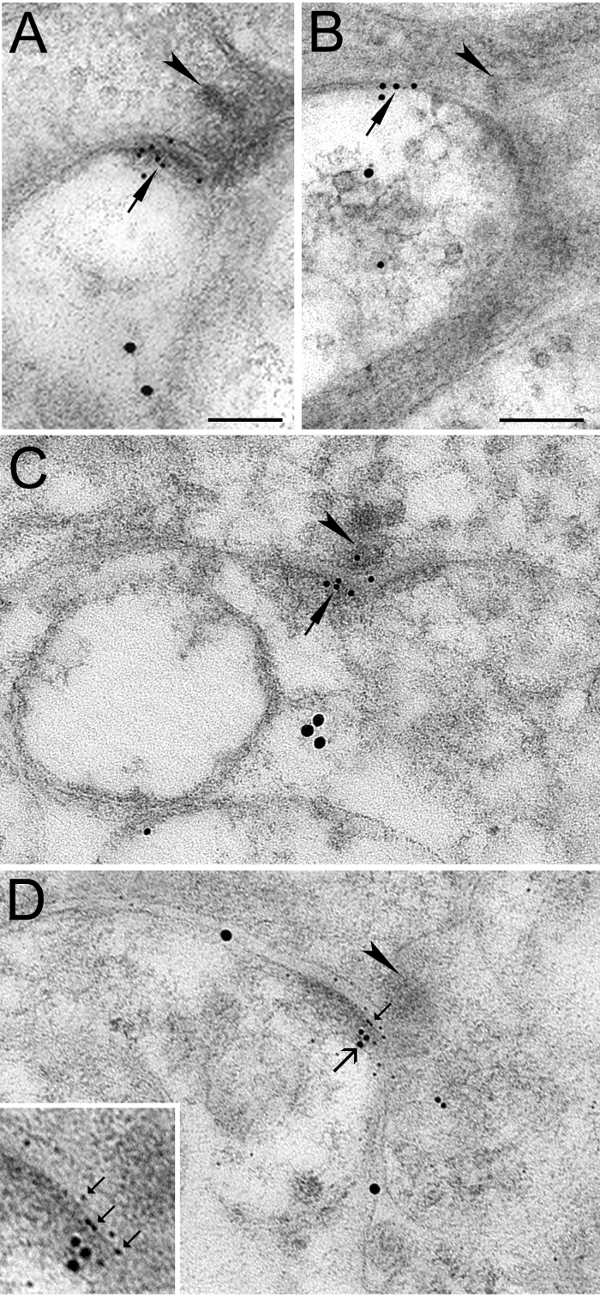
Figure 1. Immunogold Labeling Showing Synaptic and Perisynaptic Localization of Glutamate Receptors at RGC Dendrites Labeled by CTB. (A): double immunogold labeling of GluA2/3 (10 nm gold) and CTB (18 nm gold). Presynaptic ribbons indicated by arrowheads. Small gold particles are clustered (arrow) in the PSD of RGC processes. (B) double immunogold labeling of GluN2B (10 nm gold) and CTB (18 nm gold); small gold particles (arrow) are on the extrasynaptic plasma membrane. (C) double immunogold labeling of GluN2A (10 nm gold) and CTB (18 nm gold). Similar to GluA2/3, small particles are clustered within the PSD. (D) Triple immunogold labeling of GluN2A (5 nm gold), PSD-95 (10 nm gold) and CTB (18 nm gold). GluN2A gold particles (small arrows) and PSD-95 gold particles (large arrows) are co-localized within the PSD on individual CTB-positive RGC dendrites. Inset: higher magnification of the PSD. Scale bar: 0.1 µm (A = C = D, B). These are new micrographs based on data published in Zhang and Diamond 24,25. Please click here to view a larger version of this figure.
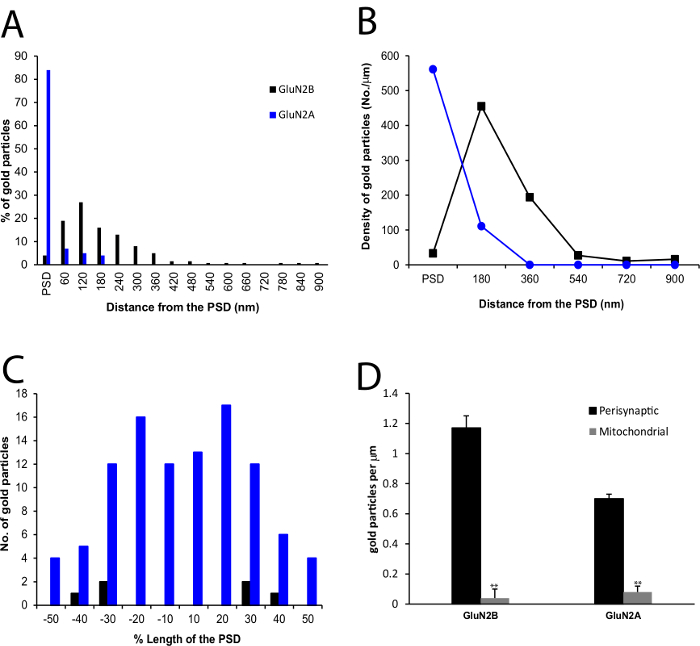
Figure 2. Quantitative Comparison of NMDAR Localization in RGC Dendrites. (A) A histogram showing the tangential distribution of immunogold for GluN2A (n = 53 profiles ) and GluN2B (n = 56 profiles ) within and outside the PSD. The perisynaptic region was divided into 60 nm bins. (B) A histogram showing labeling density of immunogold particles for GluN2A (n = 53 profiles) and GluN2B (n = 56 profiles). The perisynaptic region was divided into 180 nm bins from the edge of the PSD. (C) A histogram showing the tangential distribution of the total number of immunogold particles for GluN2A (n = 44 profiles) and GluN2B (n = 3 profiles) at all profiles with labeling within the PSD. (D) Comparison of particle density of immunogold particles for GluN2B (n = 53 and 33 profiles) and GluN2A (n = 9 and 30) in the perisynaptic membranes and mitochondrial membrane, respectively, in the CTB-positive RGC processes. These histograms are modified from histograms published in Zhang and Diamond 24,25. Please click here to view a larger version of this figure.
Discussion
We have described four techniques for successful quantitative post-embedding immunogold EM: 1) short and weak fixation, 2) freeze-substitution, 3) post-embedding immunogold staining, and 4) quantification.
EM immunogold allows the detection of specific proteins in ultrathin tissue sections. Antibodies labeled with gold particles can be directly visualized using EM. While powerful in detecting the subsynaptic localization of a membrane receptor, EM immunogold can be technically challenging, and requires rigorous optimization of tissue fixation and processing methods.
To strike an optimal balance between membrane integrity and antigenicity, we tested different fixation methods and times. For example, a successful protocol used for postembedding EM NMDAR at central synapses 19,20 failed to detect NMDAR immunoreactivity in the retina. This study employed gentle fixation and short incubation times to avoid reducing of NMDA receptor antigenicity after strong, prolonged fixation 16.
The freeze-substitution method was first developed for ultrastructural localization of glutamate receptors in the early 1990s 27, and involves low temperature infiltration and polymerization in a nonpolar, hydrophobic resin, HM20 28, needed for optimal postembedding immunogold labeling. The specific freeze-substitution method used here was developed for labeling glutamate receptors in the cochlea 29 and optimized for glutamate receptors in central synapses 18,19,20,30, and it has been modified further in recent years. The low temperature infiltration and polymerization of the resin helps to preserve antigenicity of heat-labile molecules and minimizes lipid extraction and adverse chemical reactions between resin and tissue 19,31. Combined with the fixation methods described above, this freeze-substitution method provides good antigenicity and reasonable structural preservation.
The postembedding immunogold technique used here has several advantages compared with the preembedding immunoperoxidase method. Although the preembedding immunoperoxidase method has higher sensitivity 15,16,17,19,20,21, diffusion of reaction product is unavoidable, resulting in nonspecific labeling 26,32. Moreover, the peroxidase enzyme reaction is not linear 21, making it difficult to evaluate quantitatively the precise subsynaptic localization of receptors 18,21. The postembedding immunogold method employs a non-diffusible marker and performs the immunoreaction on the surface of resin-embedded tissue, which reduces diffusible artifacts and permits higher spatial resolution 18,33,34. Gold particles can be directly counted, moreover, making it possible to estimate the number, density and variability of these receptors at retinal ribbon synapses. In addition, postembedding immunogold enables localization of multiple receptors to be simultaneously examined at the EM level. This protocol has been successfully employed in identifying localization of other membrane proteins (e.g., GABA receptors and calcium-activated potassium [BK] channels) in retinal rod bipolar synapses35.
Several factors are crucial for the success of this protocol. First, fast and gentle fixation is the first important step for the preservation of antigenicity and fine structure. Second, the "pH-shift" protocol for fixation is beneficial for optimal detection of the antigenic epitopes and good ultrastructural preservation 36. Third, freeze-substitution preserves the antigenicity of the receptors, while maintaining reasonable fine structure. Fourth, during immunogold staining, TBST buffer (with Triton X-100) is used with antibody incubation, while TBS buffer (without Triton X-100) is used for washing between and after the incubation of antibodies; this may minimize alterations in fine structure. Fifth, during exchange of solutions, sections are kept wet to reduce artifacts caused by tissue drying.
While the postembedding immunogold protocol described here provides an enhanced method for identifying membrane receptors at the retinal ribbon synapse, it is not quite as sensitive as preembedding immunohistochemical methods; but the latter are not a good option because they suffer from relatively lower specificity and are less amenable to quantification as noted above. Our protocol also compromises fine structure to some extent, as compared to more strongly fixed tissue not prepared for immunolabeling. Hopefully, future innovations will overcome these limitations.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the Intramural Programs of the National Institute of Neurological Disorders and Stroke (NINDS) and National Institute on Deafness and Other Communication Disorders (NIDCD), of the National Institutes of Health (NIH). We thank the NINDS EM facility and the NIDCD advanced imaging core (code # ZIC DC 000081-03) for assistance.
Materials
| Paraformaldehyde | EMS | 15710 | |
| Glutarldehyde | EMS | 16019 | |
| NaH2PO4 | Sigma | S9638 | |
| Na2HPO4 | Sigma | 7782-85-6 | |
| CaCl2 | Sigma | C-8106 | |
| BSA | Sigma | A-7030 | |
| Triton X-100 | Sigma | T-8787 | |
| NaOH | Sigma | 221465 | |
| NaN3 | JT Baker | V015-05 | |
| Glycerol | Gibco BRL | 15514-011 | |
| Lowicryl HM 20 | Polysciences | 15924-1 | |
| Tris-Base | Fisher | BP151-500 | |
| Tris | Fisher | 04997-100 | |
| Anti-GluN2A | Millipore | AB1555P | Dilution 1/50 |
| Anti-GluN2B | Millipore | AB1557P | Dilution 1/30 |
| Anti-GluA2/3 | Millipore | AB1506 | Dilution 1/30 |
| Anti-PSD-95 | Millipore | MA1–046 | Dilution 1/100 |
| Donkey anti-rabbit IgG-10 nm gold particles | EMS | 25704 | Dilution 1/20 |
| Donkey anti-mouse IgG-10 nm gold particles | EMS | 25814 | Dilution 1/20 |
| Donkey anti-mouse IgG-5 nm gold particles | EMS | 25812 | Dilution 1/20 |
| Donkey anti-goat IgG-18 nm gold particles | Jackson ImmunoResearch | 705-215-147 | Dilution 1/20 |
| Formvar-Carbon coated nickel-slot grids. | EMS | FCF2010-Ni | |
| Uranyl acetate | EMS | 22400-1 | |
| Methanol | EMS | 67-56-1 | |
| Lead citrate | Leica | ||
| Leica EM AFS | Leica | ||
| Leica EM CPC | Leica | ||
| Ultromicrotome | Leica | ||
| JEOL 1200 EM | JEOL | ||
| liquid nitrogen | Roberts Oxygen | ||
| Propane | Roberts Oxygen | ||
| CTB | List Biological Laboratories | 104 | 1-1.2% |
| Anti-CTB | List Biological Laboratories | 703 | Dilution 1/4000 |
Riferimenti
- Copenhagen, D. R., Jahr, C. E. Release of endogenous excitatory amino acids from turtle photoreceptors. Nature. 341, 536-539 (1989).
- Wässle, H., Boycott, B. B. Functional architecture of the mammalian retina. Physiol. Rev. 71, 447-480 (1991).
- Mittman, S., Taylor, W. R., Copenhagen, D. R. Concomitant activation of two types of glutamate receptor mediates excitation of salamander retinal ganglion cells. J. Physiol. 428, 175-197 (1990).
- Matsui, K., Hosoi, N., Tachibana, M. Excitatory synaptic transmission in the inner retina: paired recordings of bipolar cells and neurons of the ganglion cell layer. J. Neurosci. 18, 4500-4510 (1998).
- Chen, S., Diamond, J. S. Synaptically released glutamate activates extrasynaptic NMDA receptors on cells in the ganglion cell layer of rat retina. J. Neurosci. 22, 2165-2173 (2002).
- Diamond, J. S., Copenhagen, D. R. The contribution of NMDA and non-NMDA receptors to the light-evoked input-output characteristics of retinal ganglion cells. Neuron. 11, 725-738 (1993).
- Lukasiewicz, P. D., Wilson, J. A., Lawrence, J. E. AMPA-preferring receptors mediate excitatory synaptic inputs to retinal ganglion cells. J. Neurophysiol. 77, 57-64 (1997).
- Higgs, M. H., Lukasiewicz, P. D. Glutamate uptake limits synaptic excitation of retinal ganglion cells. J. Neurosci. 19, 3691-3700 (1999).
- Taylor, W. R., Chen, E., Copenhagen, D. R. Characterization of spontaneous excitatory synaptic currents in salamander retinal ganglion cells. J. Physiol. 486, 207-221 (1995).
- Kennedy, M. B. Origin of PDZ (DHR, GLGF) domains. Trends. Biochem. Sci. 20, 350 (1995).
- Kim, E., Sheng, M. PDZ domain proteins of synapses. Nat. Rev. Neurosci. 5, 771-781 (2004).
- Migaud, M., et al. Enhanced long-term potentiation and impaired learning in mice with mutant postsynaptic density-95 protein. Nature. 396, 433-439 (1998).
- Aoki, C., et al. Electron microscopic immunocytochemical detection of PSD-95, PSD-93, SAP-102, and SAP-97 at postsynaptic, presynaptic, and nonsynaptic sites of adult and neonatal rat visual cortex. Synapse. 40, 239-257 (2001).
- Davies, C., Tingley, D., Kachar, B., Wenthold, R. J., Petralia, R. S. Distribution of members of the PSD-95 family of MAGUK proteins at the synaptic region of inner and outer hair cells of the guinea pig cochlea. Synapse. 40, 258-268 (2001).
- Hartveit, E., Brandstätter, J. H., Sassoè-Pognetto, M., Laurie, D. J., Seeburg, P. H., Wässle, H. Localization and developmental expression of the NMDA receptor subunit NR2A in the mammalian retina. J. Comp. Neurol. 348, 570-582 (1994).
- Fletcher, E. L., Hack, I., Brandstätter, J. H., Wässle, H. Synaptic localization of NMDA receptor subunits in the rat retina. J. Comp. Neurol. 420, 98-112 (2000).
- Pourcho, R. G., Qin, P., Goebel, D. J. Cellular and subcellular distribution of NMDA receptor subunit NR2B in the retina. J. Comp. Neurol. 433, 75-85 (2001).
- Ottersen, O. P., Landsend, A. S. Organization of glutamate receptors at the synapse. Eur. J. Neurosci. 9, 2219-2224 (1997).
- Petralia, R. S., Wenthold, R. J., Ariano, M. A. Glutamate receptor antibodies: Production and immunocytochemistry. Receptor Localization: Laboratory Methods and Procedures. , 46-74 (1998).
- Petralia, R. S., Wenthold, R. J. Immunocytochemistry of NMDA receptors. Methods. Mol. Biol. 128, 73-92 (1999).
- Nusser, Z. AMPA and NMDA receptors: similarities and differences in their synaptic distribution. Curr. Opin. Neurobiol. 10, 337-341 (2000).
- Qin, P., Pourcho, R. G. Localization of AMPA-selective glutamate receptor subunits in the cat retina: a light- and electron-microscopic study. Vis. Neurosci. 16, 169-177 (1999).
- Zhang, J., Wang, H. H., Yang, C. Y. Synaptic organization of GABAergic amacrine cells in salamander retina. Visual. Neurosci. 21, 817-825 (2004).
- Zhang, J., Diamond, J. S. Distinct perisynaptic and synaptic localization of NMDA and AMPA receptors on ganglion cells in rat retina. J. Comp. Neurol. 498, 810-820 (2006).
- Zhang, J., Diamond, J. S. Subunit- and Pathway-Specific Localization of NMDA Receptors and Scaffolding Proteins at Ganglion Cell Synapses in Rat Retina. J. Neurosci. 29, 4274-4286 (2009).
- Peters, A., Palay, S., Webster, H. . The fine structure of the nervous system: neurons and their supporting cells, 3rd ed. , (1991).
- Baude, A., Nusser, Z., Roberts, J. D. B. The metabotropic glutamate receptor (mGluR1) is concentrated at perisynaptic membrane of neuronal subpopulations as detected by immunogold reaction. Neuron. 11, 771-787 (1993).
- Van Lookeren Campagne, M., Oestreicher, A. B., van der Krift, T. P., Gispen, W. H., Verkleij, A. J. Freeze-substitution and Lowicryl HM20 embedding of fixed rat brain: Suitability for immunogold ultrastructural localization of neural antigens. J. Hist. Cyt. 39, 1267-1279 (1991).
- Matsubara, A., Laake, J. H., Davanger, S., Usami, S., Ottersen, O. P. Organization of AMPA receptor subunits at a glutamate synapse: A quantitative immunogold analysis of hair cell synapses in the rat organ of Corti. J. Neurosci. 16, 4457-4467 (1996).
- Petralia, R. S., et al. Organization of NMDA receptors at extrasynaptic locations. Neuroscienze. 167, 68-87 (2010).
- Merighi, A., Polak, J. M., Priestley, J. V. Post-embedding electron microscopic immunocytochemistry. Electron Microscopic Immunocytochemistry: Principles and Practice. , 51-87 (1992).
- Ottersen, O. P., Takumi, Y., Matsubara, A., Landsend, A. S., Laake, J. H., Usami, S. Molecular organization of a type of peripheral glutamate synapse: the afferent synapses of hair cells in the inner ear. Prog. Neurobiol. 54, 127-148 (1998).
- Nusser, Z., Lujan, R., Laube, G., Roberts, J. D., Molnar, E., Somogyi, P. Cell type and pathway dependence of synaptic AMPA receptor number and variability in the hippocampus. Neuron. 21, 545-559 (1998).
- Takumi, Y., Ramirez-Leon, V., Laake, P., Rinvik, E., Ottersen, O. P. Different modes of expression of AMPA and NMDA receptors in hippocampal synapses. Nat. Neurosci. 2, 618-624 (1999).
- Grimes, W. N., et al. Complex inhibitory microcircuitry regulates retinal signaling near visual threshold. J. Neurophysiol. 114, 341-353 (2015).
- Sassoe-Pognetto, M., Ottersen, O. P. Organization of ionotropic glutamate receptors at dendrodendritic synapses in the rat olfactory bulb. J. Neurosci. 20, 2192-2201 (2000).

