Localization and Quantification of Begomoviruses in Whitefly Tissues by Immunofluorescence and Quantitative PCR
Summary
We describe an immunofluorescence and quantitative PCR method for the localization and quantification of begomoviruses in insect tissues. The immunofluorescence protocol can be used to colocalize viral and vector proteins. The quantitative PCR protocol can be extended to quantify viruses in whole whitefly bodies and virus-infected plants.
Abstract
Begomoviruses (genus Begomovirus, family Geminiviridae) are transmitted by whiteflies of the Bemisia tabaci complex in a persistent, circulative manner. Considering the extensive damage caused by begomoviruses to crop production worldwide, it is imperative to understand the interaction between begomoviruses and their whitefly vector. To do so, localization and quantification of the virus in the vector tissues is crucial. Here, using tomato yellow leaf curl virus (TYLCV) as an example, we describe a detailed protocol to localize begomoviruses in whitefly midguts, primary salivary glands, and ovaries by immunofluorescence. The method is based on the use of specific antibodies against a virus coat protein, dye-labeled secondary antibodies, and a confocal microscope. The protocol can also be used to colocalize begomoviral and whitefly proteins. We further describe a protocol for the quantification of TYLCV in whitefly midguts, primary salivary glands, hemolymph, and ovaries by quantitative PCR (qPCR). Using primers specifically designed for TYLCV, the protocols for quantification allow the comparison of the amount of TYLCV in different tissues of the whitefly. The described protocol is potentially useful for the quantification of begomoviruses in the body of a whitefly and a virus-infected plant. These protocols can be used to analyze the circulation pathway of begomoviruses in the whitefly or as a complement to other methods to study whitefly-begomovirus interactions.
Introduction
In the last decades, begomoviruses (genus Begomovirus, family Geminiviridae) have caused serious damage to the production of many vegetable, fiber, and ornamental crops worldwide1. Begomoviruses are transmitted in a persistent manner by the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae), which is a complex species containing over 35 cryptic species2,3. Begomoviruses may directly or indirectly affect whitefly physiology and behavior, such as fecundity4, longevity4, and host preference5,6. Furthermore, the transmission efficiency of a given begomovirus species/strain varies for different whitefly cryptic species even under the same experimental conditions7,8,9,10, indicating that there is a complex interaction between begomoviruses and whiteflies. To better understand the mechanisms underlying whitefly-begomovirus interactions, localization and quantification of the virus in whitefly tissues are essential.
Tomato yellow leaf curl virus (TYLCV) is a begomovirus that was first reported in Israel but nowadays causes serious damage to tomato production worldwide11,12. Due to its economic importance, it is one of the best-studied begomoviruses13. Like other monopartite begomoviruses, TYLCV is a single-strand circular DNA virus with a genome size of about 2,800 nucleotides14. While still under debate, several lines of evidence support the replication of TYLCV in whiteflies15,16,17. Moreover, the interaction of TYLCV particles and whitefly proteins has been reported6,18,19,20. For virus transmission, whiteflies acquire TYLCV by feeding on virus-infected plants, virions pass along the food canal to reach the esophagus, penetrate the midgut wall to reach the hemolymph, and then translocate into the primary salivary glands (PSGs). Finally, virions are egested with saliva along the salivary duct into plant phloem21. Furthermore, several studies show that TYLCV is able to be transovarially transmitted from female whiteflies to their offspring22,23. In other words, to achieve productive transmission, the virus has to overcome cellular barriers within the whitefly to translocate from one tissue to another. During the crossing of these barriers, interactions between whitefly and virus proteins are likely to occur, probably determining the efficiency with which the viruses are transmitted.
Immunofluorescence is a commonly used technique for protein distribution analysis. The specificity of antibodies binding to their antigen form the basis of immunofluorescence. Due to the economic significance of TYLCV, monoclonal antibodies against the TYLCV coat protein have been developed, offering a highly sensitive way to localize the virus24. Quantitative PCR (qPCR) allows sensitive and specific quantification of nucleic acids. This technique is most frequently based on the use of hydrolysis probe (e.g., TaqMan) or fluorescent dye (e.g., SYBR Green) detection. For hydrolysis probe-based qPCR, specific probes are needed, which consequently increase the cost. Fluorescent dye-based qPCR is simpler and more cost-effective, because labeled amplicon-specific hybridization probes are not required25. So far, several studies have used immunofluorescence and qPCR along with other methods to investigate the complex begomovirus-whitefly interactions. For example, Pan et al. performed qPCR and immunofluorescence analysis of the virus in whitefly tissues and found that the difference in ability to transmit tobacco curly shoot virus (TbCSV) between whitefly species AsiaII 1 and Middle East Asia Minor 1 (MEAM1) was due to the virus being able to efficiently cross the midgut wall of AsiaII1 but not MEAM18. Similarly, while Mediterranean (MED) whiteflies can readily transmit TYLCV, they fail to transmit tomato yellow leaf curl China virus (TYLCCNV). Selective transmission was investigated using immunofluorescence detection of virus in the PSGs, which showed that TYLCCNV do not easily cross the PSGs of MED whiteflies26. Immunofluorescence colocalization of TYLCV CP and the autophagy marker protein ATG8-II in whitefly midguts shows that autophagy plays a critical role in repressing the infection of TYLCV in the whitefly27.
Here, using TYLCV as an example, we describe a protocol for the localization of begomoviruses in whitefly midguts, PSGs, and ovaries by an immunofluorescence technique. The technique includes dissection, fixation, and incubation with primary and dye-labeled secondary antibodies. Fluorescence signals showing the location of viral proteins in the whitefly tissues can then be detected under a confocal microscope. More importantly, this protocol can be used to colocalize begomoviral and whitefly proteins. We further describe a protocol for the quantification of TYLCV using SYBR Green-based qPCR in whitefly midguts, PSGs, hemolymph, and ovaries, that can be used to compare the amount of virus in different whitefly tissue samples.
Protocol
1. Whitefly, Virus, Plants, and Acquisition of the Virus
- Rear whiteflies (MEAM1) on cotton (Gossypium hirsutum cv. Zhemian 1793) in insect-proof cages in a greenhouse at 26 ± 1 °C with a 14:10 light:dark cycle and 60 ± 10% relative humidity.
- Perform conventional PCR based on whitefly mitochondrial cytochrome oxidase I gene to determine the purity of the whitefly population.
- Collect 20 adult whiteflies and transfer them individually to a PCR tube containing 30 μL of lysis buffer (10 mM Tris, pH = 8.4, 50 mM KCl, 0.45% [wt/vol] Tween-20, 0.2% [wt/vol] gelatin, 0.45% [vol/vol] Nonidet P 40, 60 g/mL Proteinase K).
- Add 5–7 ceramic beads (2 mm in diameter) into each PCR tube and grind the samples to perform the tissue lysis.
- Incubate all samples at 65 °C for 1 h followed by 10 min at 100 °C, and then centrifuge briefly. Use this supernatant as the template for PCR amplification.
NOTE: The incubation time at 65 °C can be increased if necessary. - Prepare the PCR reaction as follows: 1 U of Taq DNA polymerase, 2 µL of 10x buffer (with Mg2+), 1.6 µL of dNTP mixture (2.5 mM), 0.5 µL of each primer (10 μM each), 2 µL of whitefly tissue lysate supernatant, and double distilled water to a total volume of 20 µL per reaction. The forward primer sequence is 5'-TTGATTTTTTGGTCATCCAGAAGT-3' and the reverse primer sequence is 5'-TAATATGGCAGATTAGTGCATTGGA-3'.
NOTE: A negative control where the DNA is substituted with nuclease-free water and a positive control with previously verified MEAM1 whitefly DNA should be included. - Perform PCR using the following cycling parameters: initial denaturation at 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 30 s, annealing at 50–55 °C for 30 s, and extension at 72 °C for 1 min, followed by 72 °C for 10 min.
- Digest the amplified product with restriction enzyme Taq I. To do so, prepare the digestion mixture with 0.1 µL (1 U) of enzyme,1 μL of 10x Taq I Buffer, 1 μL of BSA, 5 μL of amplified product, and double distilled water to a total volume of 10 μL. Incubate at 65 °C for 1 h in a thermocycler.
- Perform agarose gel electrophoresis of the samples by loading 5–7 µL of each digested product and a DNA ladder (100-2,000 bp) into the wells of a 1% agarose gel. Then, observe the DNA band sizes by gel-red staining in a gel documentation machine using UV light. Compare the band sizes of the digested products to the positive control to determine the purity of the whitefly population.
- Agro-inoculate the infectious clone of TYLCV (GenBank accession number AM282874.1) (pBINPLUS-SH2-1.4A) into 3–4 true leaf stage tomato plants (Solanum lycopersicum L. cv. Hezuo 903).
- Inoculate 10 mL of Luria–Bertani (LB) medium containing kanamycin (50 μg/mL) and rifampicin (50 μg/mL) with a single colony. Incubate at 28 °C with shaking at 200 rpm until the OD600 reaches ~1.5.
- Harvest agrobacteria by centrifugation at 4,000 x g for 10 min. Discard the supernatant.
- Resuspend pellets in 10 mL of infiltration buffer, consisting of 200 μM acetosyringone, 10 mM 2-(N-morpholino) ethane sulfonic acid (MES), and 10 mM MgCl2. Incubate at room temperature (RT) for 1–2 h.
- Infiltrate 2–3 plant leaves using the mixture from step 1.3.3 with a 1 mL needleless syringe. Maintain plants in a greenhouse at 26 ± 1 °C.
- About 1 month later, perform a visual inspection and choose plants showing yellow curl leaf and stunt symptoms.
- Transfer non-viruliferous whitefly adults onto TYLCV-infected tomato plants for 48 h for virus acquisition.
2. Dissection and Collection of Midguts, Primary Salivary Glands (PSG), Ovaries, and Hemolymph of Female Whiteflies
- Collect whiteflies using aspiration, and then place them on ice to put them in a coma. Add 1x PBS (phosphate-buffered saline) buffer onto a microscope slide, and then place whiteflies in the 1x PBS buffer for dissection under a stereo microscope.
- For the midgut dissection, break the abdomen of the whitefly and pull out the midgut using a fine acupuncture needle (0.25 mm in diameter). Transfer the midgut to clean, 1x PBS.
- For PSG dissection, break the body of the whitefly in the thorax near the head from the dorsal side. Next, insert a needle into the thorax to fix the whitefly, and use another needle to shake the whitefly until the two salivary glands are separated from the body. Then, transfer the PSGs to clean, 1x PBS.
- For the ovary dissection, completely break the abdomen of the whitefly, and use a needle to separate the ovaries from other tissues. Then, remove excess tissues by pipetting.
- For the hemolymph collection, add 10 μL of 1x PBS buffer onto a microscope slide, and then place a whitefly in the buffer. To obtain the hemolymph, gently tear a hole in the abdomen using a fine acupuncture needle, and slightly press the abdomen to release the hemolymph out into the buffer. Collect 8 μL of the liquid as the hemolymph sample.
3. Localization of TYLCV in Whitefly Midgut, Primary Salivary Glands, and Ovaries by Immunofluorescence
NOTE: The procedure for the localization of TYLCV in the midguts is presented as an example. The method can be used for PSGs and ovaries.
- Dissect the midguts in TBS buffer (10 mM Tris-HCl, 150 mM sodium chloride, pH = 7.5) as described in step 2.2.1. Collect 15–20 whitefly midguts, transfer them into a glass Petri dish (3.5 cm in diameter), and then remove excess TBS buffer.
- Add 1 mL of 4% paraformaldehyde to fix midguts for 2 h at room temperature, and then remove the fixative solution. Pipette slowly to wash the midguts 3x in TBS for 2 min each.
- Incubate the midguts in 1 mL of 0.5% Triton X-100 for 30 min to permeabilize. Then remove the permeabilization solution and wash the midguts 3x with TBST (1x TBS with 0.05% Tween 20) as described in step 3.2.
- Add 1 mL of 1% BSA (bovine serum albumin prepared in TBST) to block the midguts. Perform the blocking step at RT for 2 h, and then remove the blocking solution and wash the midguts 3x in TBST.
- Dilute the primary antibodies (MAb 1C4 against TYLCV coat protein) at 1:400 with TBST24 and add the solution into the Petri dish to immerse the midguts. Incubate at 4 °C overnight in the dark. Discard the primary antibody solution and wash 3x in TBST.
- Dilute the secondary antibodies (anti-mouse) at 1:400 with TBST and add the solution into the Petri dish to immerse the midguts. Incubate the dish for 2 h at RT in the dark. Discard the secondary antibody solution and wash the midguts 3x in TBST.
- Transfer the midguts to a clear microscope slide. Aspirate as much TBST off the slide as possible. Add 1 drop of DAPI (4', 6-diamidino-2-phenylindole, dihydrochloride) to stain the nucleus, and then place the coverslip and seal with nail polish. Examine under a confocal microscope.
4. Quantification of TYLCV in Whitefly Midgut, Primary Salivary Glands, Hemolymph, and Ovaries with qPCR
- Dissect the whitefly midguts, PSGs, hemolymph, and ovaries in 1x PBS as described above.
- Extract the DNA of the midguts, PSGs, hemolymph, and ovaries.
- After dissection, wash the midguts, PSGs, and ovaries in clean, 1x PBS buffer 2–3x.
- Collect 20 midguts or 20 PSGs in 5 μL of 1x PBS and transfer them into 25 μL lysis solution. Transfer each ovary in 5 μL of 1x PBS and then transfer into 25 μL lysis solution individually.
- Grind midguts, PSGs, and ovaries samples as described in step 1.2.2.
- Collect 8 μL of hemolymph with 1x PBS into a PCR tube, and then add 10 μL of lysis solution. Mix thoroughly.
- Extract DNA from all samples as described in 1.2.3.
- Prepare two separate qPCR master mixes (18 μL each) for TYLCV and the internal control (β-actin gene) as follows: 10 μL of SYBR Green mix, 0.8 μL of each primer (10 μM each), and 6.4 μL of double distilled water per reaction. For TYLCV, the forward primer sequence is 5′-GAAGCGACCAGGCGATATAA-3′ and the reverse primer sequence is 5′-GGAACATCAGGGCTTCGATA-3′. For β-actin, the forward primer sequence is 5′-TCTTCCAGCCATCCTTCTTG-3′ and the reverse primer sequence is 5′-CGGTGATTTCCTTCTGCATT-3′.
- Dispense 18 µL of the master mix into vials of qPCR strip tubes or a 96 well plate. Then, add 2 µL of the DNA samples into each vial. Perform all reactions with at least three technical replicates and three biological replicates.
NOTE: Step 4.5 should be performed on ice. - Close the qPCR tubes or seal the 96 well plates with adhesive plate seal.
- Centrifuge to collect the liquid at the bottom of the qPCR tubes or 96 well plate.
- Place the tubes or plate into the real-time thermal cycler and perform qPCR.
- Perform the qPCR using the following cycling parameters: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s and 60 °C for 34 s for primer annealing and elongation, 1 cycle at 95 °C for 15 s, ending with a melting curve step of 60 °C to 95 °C with an increment of 0.5 °C per 15 s.
- Choose SYBR as the fluorophore dye and unknown as sample type.
- Export the quantitative data to a spreadsheet format and analyze the relative quantity of viral DNA by the 2−ΔΔCT method28.
Representative Results
The MEAM1 whiteflies of the B. tabaci complex and TYLCV were used as an example here to describe the procedures. The overview of the immunofluorescence and viral quantification procedures described in this manuscript is shown in Figure 1. Figure 2 shows representative results of immunofluorescence detection of TYLCV and DAPI staining in PSGs, midguts, and ovaries, indicating that TYLCV accumulated more in the PSGs and midguts, and less in the ovaries. Figure 3 shows the relative quantity of virus in the whitefly PSGs, midguts, hemolymph, and ovaries after whiteflies fed on TYLCV-infected tomato for 24, 48, and 72 h, indicating that the quantity of virus increased in different whitefly tissues with the increase of the acquisition access period.
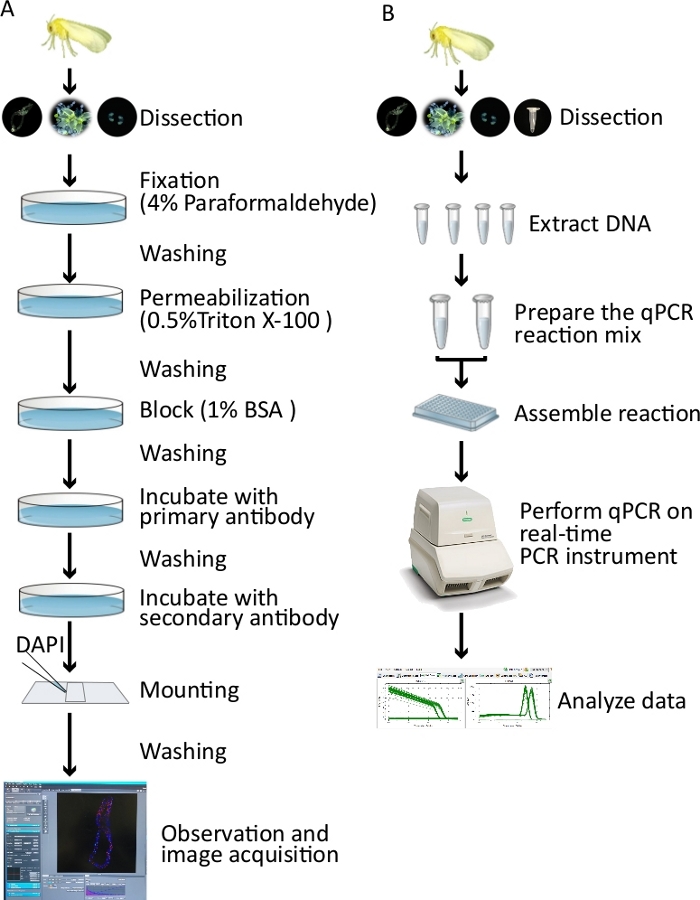
Figure 1: Overview of the protocols presented in this paper. (A) Experimental procedures for immunofluorescence in whitefly midguts, PSGs, and ovaries. Tissues were collected from TYLCV-infected adult female whiteflies, followed by a series of fixation, permeabilization, blocking, incubations with primary antibodies that bind to a TYLCV coat protein, and fluorochrome-conjugated secondary antibodies that bind to the primary antibodies. Finally, samples were analyzed under a confocal microscope, allowing the acquisition of images. (B) Experimental procedure for the quantification of virus in the whitefly midguts, PSGs, ovaries, and hemolymph. Tissues were collected from TYLCV-infected adult female whiteflies, and then DNA extraction was performed. qPCR was performed on a real-time PCR instrument, allowing the acquisition of data that can be subsequently analyzed. Please click here to view a larger version of this figure.
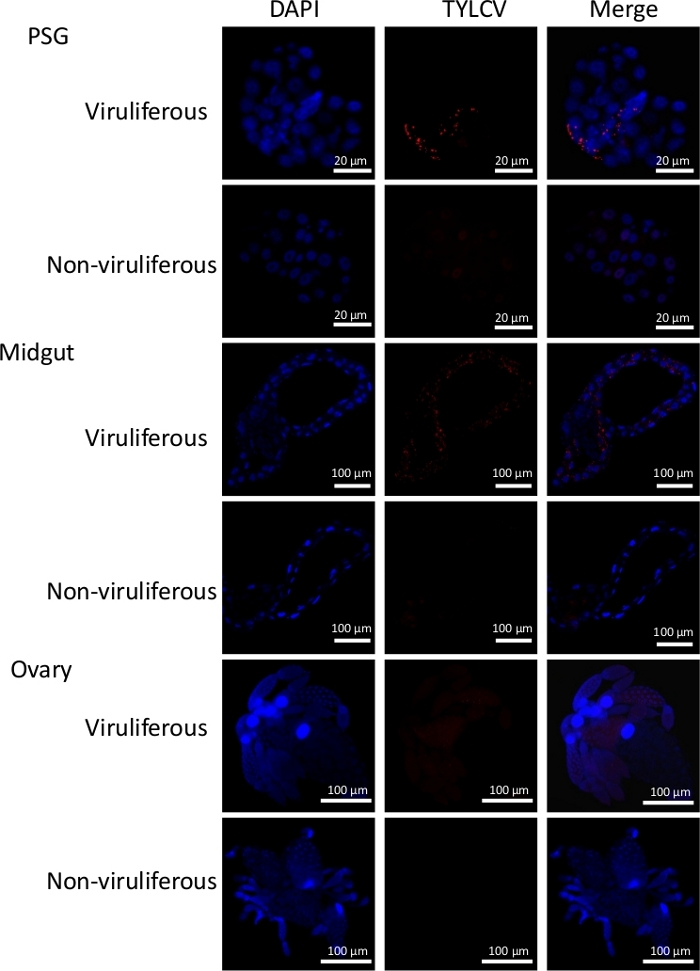
Figure 2: Immunofluorescence detection of TYLCV in the primary salivary glands, midguts, and ovaries of female adult whiteflies after a 48 h virus acquisition access period. Whiteflies were allowed to feed on TYLCV-infected tomato plants for 48 h, and then 20 PSGs, 20 midguts, and 20 ovaries were dissected and collected. Next, samples were fixed in 4% formaldehyde, permeabilized in 0.5% Triton X-100, blocked in 1% BSA, and then incubated with mouse anti-TYLCV monoclonal antibodies (MAb 1C4 against TYLCV coat protein)24 and goat anti-mouse secondary antibodies conjugated to a red fluorescent dye (i.e., Dylight 549). Nuclei were stained with DAPI (blue). Immunofluorescence signals were examined under a confocal microscope. Please click here to view a larger version of this figure.
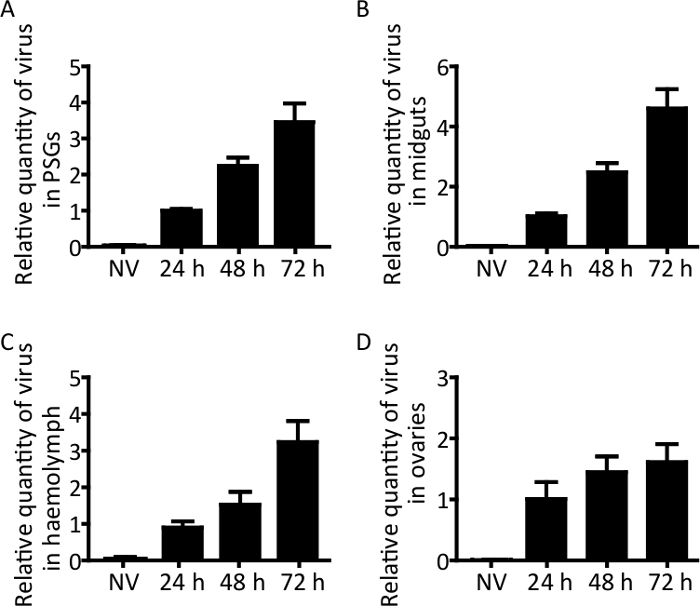
Figure 3: Quantification of TYLCV DNA in the midguts, hemolymph, primary salivary glands, and ovaries in MEAM1 whiteflies. Adult fed on TYLCV-infected tomato plants for 24, 48, and 72 h, and then the whitefly midguts, hemolymph, PSGs, and ovaries were dissected and collected. Next, extraction of DNA and qPCR were performed for the midguts (A), hemolymph (B), PSGs (C), and ovaries (D). Data were firstly normalized to the whitefly β-actin DNA, and then the relative quantity of virus obtained were again normalized to that at 24 h. NV: Non-viruliferous. Data are presented as the mean ± SEM of relative quantity of virus. Please click here to view a larger version of this figure.
Discussion
Here we describe a protocol for the localization and quantification of a begomovirus in the tissues of its whitefly vector by immunofluorescence and qPCR. Dissection represents the first step to localize and quantify the virus in whitefly tissues. The whitefly body is about 1 mm in length, which means that the tissues are extremely small, and it is difficult to dissect them. Besides, there are strong connections between the tissues. For example, the ovaries are tightly connected with the bacteriocytes, making them difficult to isolate. Here, considering the characteristics and locations of the different tissues, we describe different approaches for dissection. However, a lot of practice is needed to rapidly and accurately dissect these tissues. In addition, it is best to use fresh whiteflies, because dead insects would increase the difficulty of dissection. Furthermore, postdissection, tissues must be cleaned to avoid contamination.
The immunofluorescence technique is based on the use of specific antibodies. A suitable working concentration for each antibody is very important in imaging, because a high concentration of antibodies reduces the signal-to-background ratio and a low concentration may not provide sufficient signal. Here, we used a dilution of 1:400 for both primary antibodies and second antibodies. These antibodies should be divided into small aliquots and kept at -20 °C to avoid repeated freezing and thawing. The antibody dilutions should be prepared right before use. When the protocol described here is used to colocalize viral and whitefly proteins, it is important that the primary antibodies against viral and whitefly proteins are produced in animals of different species. Additionally, the fluorophores of the secondary antibodies should be chosen carefully to prevent bleed-through between channels. Furthermore, in view of the small size of whitefly tissues, they should always be washed under the microscope to avoid loss. Although immunofluorescence is highly sensitive, it is relatively more time-consuming and costly when compared to other virus location methods such as Fluorescence in situ Hybridizations (FISH). In addition, antibodies against begomoviruses may not be available for purchase.
The key to the successful application of qPCR for the quantification of the virus in whitefly tissues lies in the design of suitable primers and selection of endogenous reference genes. Rodríguez et al. described the criteria for designing qPCR primers in detail25, which also applies in the design of virus primers. In this study, β-actin was selected as the reference gene for virus quantification in whitefly tissues. However, in some cases (e.g., RNA genes), other endogenous genes should be used as reference genes if they are suitable for the experimental design29. Also, to obtain stable and accurate results, whiteflies of same developmental stage should be used, because whiteflies of different developmental stages possess differential capabilities to acquire and transmit viruses22. Moreover, while we used 20 PSGs or midguts as one sample, we used a hemolymph collected from one whitefly as one sample for qPCR analysis. This is due to the fact that the collection of the hemolymph is time-consuming and the DNA of the hemolymph samples is vulnerable to degradation if not processed in a timely manner. This protocol can also be used to quantify the amount of virus in whitefly whole body and virus-infected plants.
In summary, we describe an efficient and sensitive protocol to localize and quantify begomoviruses in whitefly tissues by immunofluorescence and qPCR, respectively. This protocol can also be adapted for the localization and quantification of other begomoviruses. Furthermore, the protocol of immunofluorescence can be used to colocalize viral and whitefly proteins.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by National Key Research and Development Program (Grant number: 2017YFD0200600), the earmarked fund for China Agriculture Research System (grant number: CARS-23-D07) and the Bill & Melinda Gates Foundation (Investment ID OPP1149777). We thank Prof. Jian-Xiang Wu for providing TYLCV CP antibodies.
Materials
| 4% Paraformaldehyde | MultiSciences | F0001 | |
| 4',6-diamidino-2-phénylindole (DAPI) | Abcam | ab104139 | |
| Bovine Serum Albumin (BSA) | MultiSciences | A3828 | |
| CFX Connect Real-Time PCR Detection System | Bio-RAD | 185-5201 | |
| Confocal microscopy | Zeiss | LSM800 | |
| Dylight 549-goat anti-mouse | Earthox | E032310-02 | Secondary antibody |
| Monoclonal antibody (MAb 1C4) | Primary antibody | ||
| Phosphate Buffered Saline (PBS) | Sangon Biotech | B548119-0500 | |
| Stereo microscope | Zeiss | Stemi 2000-C | |
| TB green premix Ex Taq (Tli RNase H Plus) | TaKaRa | RR820A | qPCR master mix |
| Thermocycler | Thermofisher | A41182 | |
| Tissuelyzer | Shaghai jingxin | Tissuelyser-48 | |
| Triton-X-100 | BBI life sciences | 9002-93-1 | |
| Tween 20 | BBI life sciences | 9005-64-5 |
Riferimenti
- Rojas, M. R., et al. World management of geminiviruses. Annual Review of Phytopathology. 56, 637-677 (2018).
- De Barro, P. J., Liu, S. S., Boykin, L. M., Dinsdale, A. B. Bemisia tabaci: a statement of species status. Annual Review of Entomology. 56, 1-19 (2011).
- Navas-Castillo, J., Fiallo-Olive, E., Sanchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annual Review of Phytopathology. 49, 219-248 (2011).
- Liu, J., et al. Viral infection of tobacco plants improves performance of Bemisia tabaci but more so for an invasive than for an indigenous biotype of the whitefly. Journal of Zhejiang University-Science B. 11 (1), 30-40 (2010).
- Legarrea, S., Barman, A., Marchant, W., Diffie, S., Srinivasan, R. Temporal effects of a begomovirus infection and host plant resistance on the preference and development of an insect vector, Bemisia tabaci, and implications for epidemics. PLoS One. 10 (11), 0142114 (2015).
- Fang, Y., et al. Tomato yellow leaf curl virus alters the host preferences of its vector Bemisia tabaci. Scientific Reports. 3, 2876 (2013).
- Guo, T., et al. Comparison of transmission of papaya leaf curl China virus among four cryptic species of the whitefly Bemisia tabaci complex. Scientific Reports. 5, 15432 (2015).
- Pan, L. L., et al. Differential efficiency of a begomovirus to cross the midgut of different species of whiteflies results in variation of virus transmission by the vectors. Science China-Life Sciences. 61 (10), 1254-1265 (2018).
- Pan, L. L., Cui, X. Y., Chen, Q. F., Wang, X. W., Liu, S. S. Cotton leaf curl disease: which whitefly is the vector. Phytopathology. 108 (10), 1172-1183 (2018).
- Fiallo-Olive, E., Pan, L. L., Liu, S. S., Navas-Castillo, J. Transmission of begomoviruses and other whitefly-borne viruses: dependence on the vector species. Phytopathology. , (2019).
- Cohen, S., Nitzany, F. E. Transmission and host range of the tomato yellow leaf curl virus. Phytopathology. 56, 1127-1131 (1966).
- Moriones, E., Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Research. 71 (1-2), 123-134 (2000).
- Ghanim, M. A review of the mechanisms and components that determine the transmission efficiency of tomato yellow leaf curl virus (Geminiviridae; Begomovirus) by its whitefly vector. Virus Research. 186, 47-54 (2014).
- Navot, N., Pichersky, E., Zeidan, M., Zamir, D., Czosnek, H. Tomato yellow leaf curl virus – a whitefly-transmitted geminivirus with a single genomic component. Virology. 185 (1), 151-161 (1991).
- Sanchez-Campos, S., et al. Tomato yellow leaf curl virus: No evidence for replication in the insect vector Bemisia tabaci. Scientific Reports. 6, 30942 (2016).
- Pakkianathan, B. C., et al. Replication of tomato yellow leaf curl virus in its whitefly vector, Bemisia tabaci. Journal of Virology. 89 (19), 9791-9803 (2015).
- Rodriguez-Negrete, E. A., et al. A sensitive method for the quantification of virion-sense and complementary-sense DNA strands of circular single-stranded DNA viruses. Scientific Reports. 4, 6438 (2014).
- Gotz, M., et al. Implication of Bemisia tabaci heat shock protein 70 in Begomovirus-whitefly interactions. Journal of Virology. 86 (24), 13241-13252 (2012).
- Zhao, J., Chi, Y., Zhang, X. J., Wang, X. W., Liu, S. S. Implication of whitefly vesicle associated membrane protein-associated protein B in the transmission of Tomato yellow leaf curl virus. Virology. 535, 210-217 (2019).
- Maluta, N. K., Garzo, E., Moreno, A., Lopes, J. R., Fereres, A. Tomato yellow leaf curl virus benefits population growth of the Q biotype of Bemisia tabaci (Gennadius) (Hemiptera: Aleyrodidae). Neotropical Entomology. 43 (4), 385-392 (2014).
- Czosnek, H., Hariton-Shalev, A., Sobol, I., Gorovits, R., Ghanim, M. The incredible journey of begomoviruses in their whitefly vector. Viruses. 9 (10), 273 (2017).
- Wei, J., et al. Vector development and vitellogenin determine the transovarial transmission of begomoviruses. Proceedings of the National Academy of Sciences of the United States of America. 114 (26), 6746-6751 (2017).
- Ghanim, M., Morin, S., Zeidan, M., Czosnek, H. Evidence for transovarial transmission of tomato yellow leaf curl virus by its vector, the whitefly Bemisia tabaci. Virology. 240 (2), 295-303 (1998).
- Xie, Y., et al. Highly sensitive serological methods for detecting tomato yellow leaf curl virus in tomato plants and whiteflies. Virology Journal. 10, 142 (2013).
- Arya, M., et al. Basic principles of real-time quantitative PCR. Expert Review of Molecular Diagnostics. 5 (2), 209-219 (2005).
- Wei, J., et al. Specific cells in the primary salivary glands of the whitefly Bemisia tabaci control retention and transmission of begomoviruses. Journal of Virology. 88 (22), 13460-13468 (2014).
- Wang, L. L., et al. The autophagy pathway participates in resistance to tomato yellow leaf curl virus infection in whiteflies. Autophagy. 12 (9), 1560-1574 (2016).
- Arocho, A., Chen, B. Y., Ladanyi, M., Pan, Q. L. Validation of the 2(-Delta Delta Ct) calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagnostic Molecular Pathology. 15 (1), 56-61 (2006).
- Li, R., et al. Reference gene selection for qRT-PCR analysis in the sweetpotato whitefly, Bemisia tabaci (Hemiptera: Aleyrodidae). PLoS One. 8 (1), 53006 (2013).

