Applications of RNA Interference in American Cockroach
Summary
The present protocol describes step-by-step guidelines for the RNAi operation techniques in P. americana.
Abstract
Cockroaches, a sanitary pest, are essential species in insect developmental and metamorphic studies due to their easy feeding and hemimetabolous characteristics. Altogether with well-annotated genome sequences, these advantages have made American cockroach, Periplaneta americana, an important hemimetabolous insect model. Limited by the shortage of knockout strategy, effective RNA interference (RNAi)-based gene knockdown becomes an indispensable technique in functional gene research of P. americana. The present protocol describes the RNAi operation techniques in P. americana. The protocol includes (1) selection of the P. americana at proper developmental stages, (2) preparation for the injection setting, (3) dsRNA injection, and (4) gene knockdown efficiency detection. RNAi is a powerful reverse genetic tool in P. americana. The majority of P. americana tissues are sensitive to extracellular dsRNA. Its simplicity allows researchers to quickly obtain dysfunctional phenotypes under one or multiple targeting dsRNA injections, enabling researchers to better use the P. americana for developmental and metamorphic studies.
Introduction
RNA interference (RNAi), an evolutionarily conserved mechanism, gradually becomes an essential reverse-genetic tool to inhibit gene expression in many organisms1, since Andrew Fire and Craig Mello2 developed the double-stranded RNA (dsRNA) mediated gene silence strategy. dsRNA is cleaved into fragments of 21-23 nucleotides, small interfering RNAs (siRNAs), by the enzyme Dicer in cells to activate the RNAi pathway. Then siRNAs are incorporated into the RNA-induced silencing complex (RISC), which couples to the target mRNA, causes mRNA cleavage, and finally results in the loss of gene function3,4,5. Among the insect species, many systemic RNAi experiments have so far been reported in lots of insect orders, such as Orthoptera, Isoptera, Hemiptera, Coleoptera, Neuroptera, Diptera, Hymenoptera, Lepidoptera, and Blattodea5,6,7,8.
Cockroaches (Blattaria) are an essential insect family in developmental and metamorphic studies with their rapid growth cycles, strong adaptability to the environment, and high developmental plasticity9. Before discovering that RNAi was compatible with cockroaches, previous research only focused on cockroach prevention and control due to a scarcity of genetic manipulation techniques in cockroaches. The cockroach ootheca's unique structure made it challenging to perform embryo injection-based gene knockout with the CRISPR-Cas9 system. Besides, most tissues in cockroaches (such as P. americana) show robust systemic RNAi response, allowing for the rapid generation of dysfunctional phenotypes by injecting one or more targeting dsRNAs9,10,11. These features made RNAi an indispensable technique in gene functional research in P. americana.
Even though the use of RNAi in functional gene research in P. americana has been reported, no detailed or step-by-step description was available. This report provides one step-by-step operational guideline for RNAi in P. americana, useful for gene function study in other cockroaches. Furthermore, this guide is not limited to Blattodea and can be applied to many other insects with minor modifications.
Protocol
The line of P. americana was initially provided by Dr. Huiling Hao. This species has been maintained with inbreeding for 30 years9.
1. Hatching and feeding of P. americana
- Collect fresh oothecae (immediately post egg-laying) of P. americana and incubate in the dark incubator at 25 °C and 60% humidity for ~25 days. Then increase the temperature and humidity to 30 °C and 75% 3 days before hatching.
- Use a sieve with 4 mm aperture to separate the hatched nymphs from oothecae.
- Keep the nymphs in cylindrical containers (12 cm in diameter and 10 cm in height) in the dark at 28 °C, 70% humidity. Brush the inside edge of the containers with vaseline to prevent cockroaches from escaping. Provide rat food, water, and shelter (egg trays). Pay attention to the activity of the nymphs and regularly clean up the feces and debris.
2. Selection of the nymphs in proper instar
- Use a glass tube to pick out the freshly molted P. americana (white in body color). Keep them in new containers and wait for the correct stage for treatment.
NOTE: After ~19 days under the above feeding conditions, the nymphs in the 3rd instars will be available for injection. The color of freshly molted cockroaches is white, and the instars are clarified by molting times.
3. Preparation of the target fragment with T7 promoters
- Using the cDNA of P. americana as a template, design paired primers to perform PCR to obtain 300-800 bp DNA fragment of the target gene9. Then clone the PCR fragment into a pTOPO vector (see Table of Materials) for sequencing.
- Using the target fragment DNA as a template, synthesize one new pair of primers with T7 promoter sequence (5'-GGATCCTAATACGACTCACTATAGG-3') at the 5' terminals and perform another round of PCR to obtain the target fragment with T7 promoters on each side9.
4. Transcription and synthesis of dsRNA in vitro
- Add 10 µL of T7 2X Buffer, 2 µL of the Enzyme Mix, T7 Express (see Table of Materials), and 2 µg of PCR product (obtained in step 3.2) in the reaction system and add up the total volume to 20 µL with ddH2O. Gently mix upside down manually and incubate at 37 °C for 30 min and 70 °C for 10 min, and then slowly cool to room temperature.
- Dilute RNase A solution (4 µg/µL) with ddH2O at a ratio of 1:200. Then add 1 µL of RQ1 RNase-Free DNase (1 U/µL) and 1 µL of diluted RNase A solution to the system (see Table of Materials).
NOTE: Now, the volume of a single system is 22 µL. - Incubate at 37 °C for 30 min. Then add 10% of the total volume (when performing N reactions simultaneously, the total volume is N x 22 µL) of Sodium acetate and three volumes of the total volume of isopropanol.
- Gently mix upside down manually, place on ice for 5 min, and centrifuge at 13,000 x g for 10 min at 4 °C.
- Remove the supernatant with a pipette, wash the residue with 75% ethanol (with 25% diethyl pyrocarbonate (DEPC) treated water), and then centrifuge at 13,000 x g for 5 min at 4 °C.
- Remove supernatant again. Air-dry the pellet for 15 min at room temperature. Use ~100 µL of DEPC water to dissolve dsRNA, then dilute the dsRNA to the final 2 µg/µL.
NOTE: The dsRNA could be stored at -80 °C for 6 months.
5. Loading dsRNA solution into the syringe
- Set up the program in the micro-injection pump (see Table of Materials) in advance to ensure that the volume of each injection is consistent. Before using the syringe, clean the syringe by filling it with DEPC water 8-10 times.
- Install the 10 µL Syringe on the micro-injection pump, start the pump, and fill the syringe with 10 µL of dsRNA solution (prepared at step 4.6). Inject 1 µL of the dsRNA into a 3rd instar nymph (see step 2) and ensure that it does not leak out to the body.
6. Injecting dsRNA to P. americana
- Anesthetize the cockroaches with an increased concentration of CO2 in a container until the cockroaches do not move anymore, and then proceed immediately with the injection.
- Gently pick up the cockroach with tweezers, and deliver the cockroach toward the needle with hand. Next, insert the needle via the gap between two abdominal somites horizontally against the epidermis. About insert depth, the shallower, the better.
- Then, inject the dsRNA solution into the cockroach. Finally, pull out the needle tip. Ensure that the needle tip is as close as possible to the epidermis to avoid damaging internal organs.
NOTE: The injection should be made under a dissection microscope. - Put the injected cockroaches into clean bioassay containers. Wait for about 10-20 min to let them recover from the CO2 effects. Label the containers with the date of injection, type and dose of dsRNA, and age of the P. americana.
- Place the injected cockroaches in a dark environment at 28 °C, 70% humidity, provide water, feed, and shelter, and observe possible changes in the phenotype of the cockroaches regularly.
7. Knockdown confirmation and phenotypic analysis
- Evaluate the efficiency of RNAi using any available molecular biology techniques such as quantitative Real-Time PCR (qRT-PCR) and Western blotting. For detailed qRT-PCR and Western blotting procedures, see References1,12.
- To observe and analyze RNAi-related phenotypes, use the microscope suitable for living animals.
NOTE: RNAi may affect the morphology, behavior, molting, limb regeneration, and other physiological activities of cockroaches. The specific frequency of phenotype is calculated according to specific conditions.
Representative Results
Figure 1 shows a successful injection. The microinjection syringe with a micro diameter needle should be horizontally placed on the booster (Figure 1A). The needle is inserted via the gap between two abdominal somites horizontally against the epidermis (Figure 1B). Ensure that the liquid goes into the P. americana abdomen. The too steep angle of the needle will damage the internal organs (Figure 1C), and improper injection leads to leakage of the dsRNA solution out of the abdomen (Figure 1D). If any dsRNA leaks out, the animal needs to be discarded, and another injection is performed. The P. americana needs to be completely anesthetized during the injection.
Like other biological experiments, control groups are needed when RNAi treatment is performed. The treatment differences need to be confirmed caused by targeted RNAi instead of due to the off-target effects of dsRNA, the damage during the injection, or CO2 anesthesia. Here Ddc (Dopa decarboxylase) and Dpp (decapentaplegic) genes were selected as two examples to observe the phenotype of the molted cockroaches after dsRNA injection, and a negative control dsMock11 which has no target in the animals, was performed as a negative control, the RNAi efficiency was detected by qRT-PCR (Figure 2). Taking the injection of dsDdc as an example (Figure 3A,B), the P. americana with knocked down Ddc gene would have the white epidermis for a long time since the melanization is blocked (Figure 3B). In the experiment of dsDpp injections, which are necessary for limb regeneration, the RNAi disrupted the limb regeneration (Figure 4), which was reported previously9.
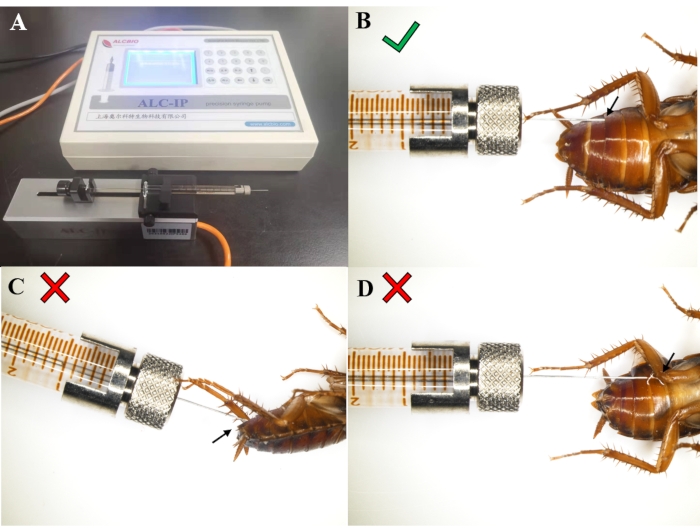
Figure 1: Instrument and correct injection operation. (A) The syringe on the microinjection instrument. (B) Correct position of the injection needle; no liquid comes out. (C) The too steep angle of the injection needle may damage the internal organs. (D) Failed injection results in the dsRNA solution or hemolymph flowing out. Please click here to view a larger version of this figure.
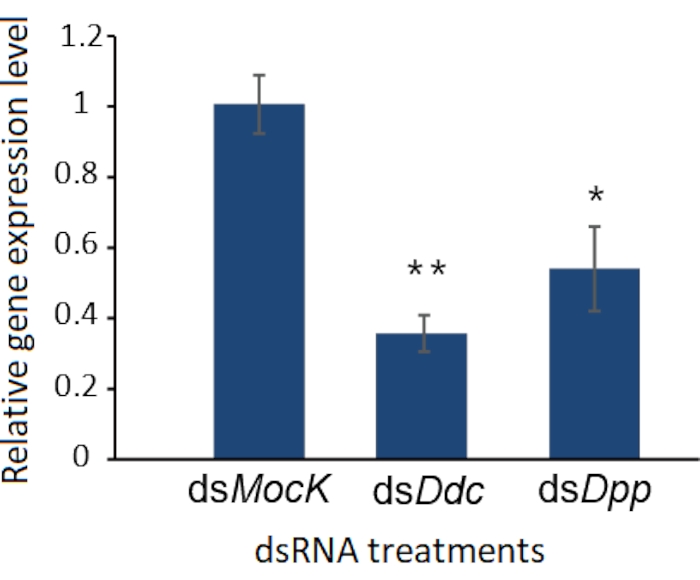
Figure 2: RNAi efficiency detected by qRT-PCR. The knockdown efficiency of Ddc and Dpp genes were detected by qRT-PCR, and the dsMock was used as a negative control. Three replicates were used for each treatment, and the error bars indicate standard deviation of the three replicates. The significance of differences was analyzed by Student's t-test. *: P < 0.05, **: P < 0.01. Please click here to view a larger version of this figure.
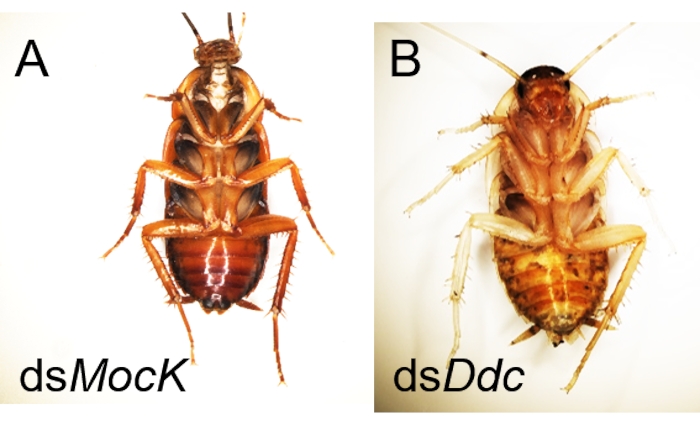
Figure 3: Examples of successful RNAi for disruption of epidermis melanization in P. americana. (A) Normal cockroach after injection ofdsMock with normal melanization. (B) Ddc RNAi mediated albinistic P. americana after molting. Please click here to view a larger version of this figure.
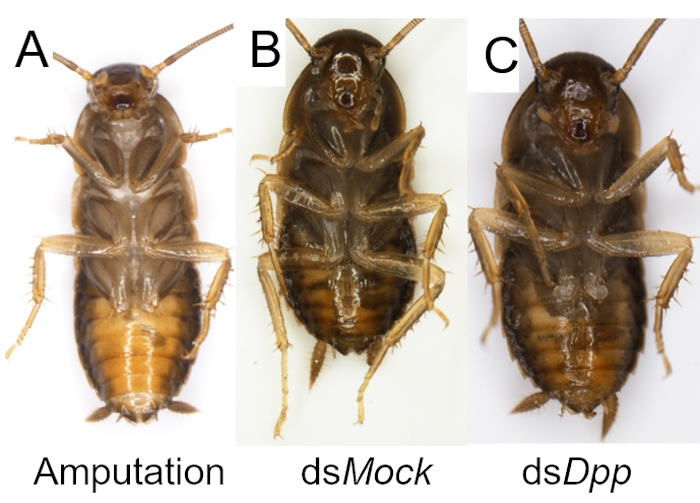
Figure 4: Examples of successful RNAi for disruption of limb regeneration in P. americana. (A) A P. americana with an amputated hind leg. (B-C) P. americana after RNAi treatments and molting. The hindleg was regenerated in dsMock control group (B) but not when the Dpp (C) gene was silenced. Please click here to view a larger version of this figure.
Discussion
This report described a methodological step-by-step RNAi strategy in P. americana; of note, it also can be applied to other cockroaches (Blattella germanica, for example) and many other insects with minor changes. However, the gene silencing efficiency of RNAi is not always high enough, with an obvious disadvantage compared with the gene knockout strategy13. The following residual effect of gene-level may interfere with the real phenotypes. To ensure the RNAi treatment is successful, several essential conditions need to be considered, including the physical condition of animals, selection of control group, the length and concentration of the dsRNA molecules, the avoiding of the possibility of Off-target effects (OTE), and different sensitivity of tissues to dsRNA.
Physical condition of P. americana
Healthy animals are the premise for the success of RNAi and different gene functional studies in P. americana. Besides, to keep experiments consistent, only cockroaches that have been raised in the same conditions are chosen. And only nymphs that are older than the 3rd instar (about 19 days post-hatching) can be used because younger nymphs are too small for injection in body size.
Selection of control group
dsMock11 and dsEGFP and the same dose of cockroach saline6 have been used as negative controls. The target sequences of negative controls should be exogenous and have no homology with the endogenous genetic sequence11. dsMock was preferred because using dsEGFP would delay the growth and development of P. americana to a certain degree.
dsRNA length
The efficiency of RNAi is affected by the length of the dsRNA molecules. Longer dsRNA fragments are more effective at mRNA silencing, possibly because it produces more siRNAs, or shorter dsRNA fragments are taken up by cells less efficiently 1. In P. americana and B. germanica, dsRNA between 300 bp and 800 bp appears ideal for RNAi experiments6,9,10,14. A short dsRNA may be insufficient to induce a systemic RNAi response, whereas a long dsRNA may increase OTE risk1.
dsRNA concentration
The concentration of dsRNA can also influence the efficiency of RNAi1,15. For the 3rd instar nymphs of P. americana, 1 µg of dsRNA appears to be a reasonable amount, and 4-6 µg is suitable for adult6,9,14,16. The precise dsRNA amount used for injection can be adjusted based on the genes, tissues, and body size of the P. americana.
Off-target effects
The OTE of RNAi is hard to avoid altogether17,18. Two non-overlapping regions of the dsRNA can be designed to reduce the effect of OTE on phenotypes5. If dsRNAs targeting two distinct regions produce the same effect, the possibility of OTE-induced false positive phenomenon could be excluded.
Method for RNAi efficiency detection
The phenotypic analysis is the most intuitive approach for evaluating RNAi, and qRT-PCR and Western blotting analysis are the two golden standards for measuring knockdown efficiency. qRT-PCR is a straightforward method for determining interference efficiency at mRNA level6,9,10. The primers should be designed out of the dsRNA targeting region. The Western blotting analysis is another method for analyzing protein-level interference but requires specific antibodies. However, no particular phenotypic effects are sometimes observed, even with high silencing efficiency (>90%). One possible explanation for this residual effect is massive gene family expansion in cockroaches; the redundancies of genes control many particular functions and phenotypes. Another possible explanation is that the minimum gene expression level required for functional enough is extremely low. These lead to failure in obtaining a particular phenotype even with high enough RNAi efficiency.
Tissue-specificity for the RNAi sensitivity
The interference efficiency in some cockroach tissues (such as the ovary and accessory glands) is not high enough, which could be the penetrating barrier for dsRNA5,19; the different RNAi efficacy in different insect species is also observed, which depends on the enzymatic degradation of dsRNA in hemolymph15,20. A few ideas are reasonable for improving RNAi efficiency in P. americana tissues, such as digesting dsRNA into siRNAs in vitro21, increasing the dsRNA dose and injection times, and using nanomaterials as a carrier for delivery.
In conclusion, the high efficiency of RNAi together with well-annotated genome sequences9 makes P. americana a good model for investigating gene function in a wide range of developmental, physiological, and behavioral studies. Similarly, this report can serve as a valuable reference for other insects sensitive to dsRNA and hard to make knockout mutations.
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant Nos. 32070500, 31620103917, 31330072, and 31572325 to C.R., Sh.L.), by the Natural Science Foundation of Guangdong Province (Grant No. 2021B1515020044 and 2020A1515011267 to C.R.), by the Department of Science and Technology in Guangdong Province (Grant Nos. 2019B090905003 and 2019A0102006), by the Department of Science and Technology in Guangzhou (Grant No. 202102020110), by the Shenzhen Science and Technology Program (Grant No. KQTD20180411143628272 to Sh.L.).
Materials
| 701 N 10 µL Syr (26s/51/2) | Hamilton | PN:80300 | Injection |
| Incubator | Ningbo Jiangnan Instrument Factory | RXZ-380A-LED | For cockroaches hatching and feeding |
| Micro-injection pump | Alcott Biotechnology | ALC-IP600 | Injection |
| pTOPO-Blunt Cloning Kit | Aidlab Biotechnology | CV16 | For Gene clonging |
| quantitative Real-Time PCR Systems | Bio-Rad | CFX Connect | For qRT-PCR analysis |
| T7 RiboMAX Express RNAi System | Promega | P1700 | For dsRNA synthesis, which contains Rnase A Solution (4 μg/μL), Sodium Acetate, 3.0M (pH 5.2), Enzyme Mix, T7 Express, Nuclease-Free water, Express T7 2x Buffer, RQ1 RNase-Free DNase |
| Thermal Cyclers | Bio-Rad | S1000 | For DNA amplification |
Riferimenti
- Miller, S. C., Miyata, K., Brown, S. J., Tomoyasu, Y. Dissecting systemic RNA interference in the red flour beetle Tribolium castaneum: Parameters affecting the efficiency of RNAi. PloS One. 7 (10), 47431 (2012).
- Fire, A., et al. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 391 (6669), 806-811 (1998).
- Ambesajir, A., Kaushik, A., Kaushik, J. J., Petros, S. T. RNA interference: A futuristic tool and its therapeutic applications. Saudi Journal of Biological Sciences. 19 (4), 395-403 (2012).
- Younis, A., Siddique, M. I., Kim, C. K., Lim, K. B. RNA interference (RNAi) induced gene silencing: A promising approach of hi-tech plant breeding. International Journal of Biological Sciences. 10 (10), 1150-1158 (2014).
- Bellés, X. Beyond Drosophila: RNAi in vivo and functional genomics in insects. Annual Review of Entomology. 55, 111-128 (2010).
- French, A. S., Meisner, S., Liu, H., Weckström, M., Torkkeli, P. H. Transcriptome analysis and RNA interference of cockroach phototransduction indicate three opsins and suggest a major role for TRPL channels. Frontiers in Physiology. 6, 207 (2015).
- Hennenfent, A., Liu, H., Torkkeli, P. H., French, A. S. RNA interference supports a role for Nanchung-Inactive in mechanotransduction by the cockroach, Periplaneta americana, tactile spine. Invertebrate Neuroscience: IN. 20 (1), 1 (2020).
- Immonen, E. V., et al. EAG channels expressed in microvillar photoreceptors are unsuited to diurnal vision. The Journal of Physiology. 595 (16), 5465-5479 (2017).
- Li, S., et al. The genomic and functional landscapes of developmental plasticity in the American cockroach. Nature Communications. 9 (1), 1008 (2018).
- Zhao, Z., et al. Grainy head signaling regulates epithelium development and ecdysis in Blattella germanica. Insect Science. 28 (2), 485-494 (2021).
- Lozano, J., Belles, X. Conserved repressive function of Krüppel homolog 1 on insect metamorphosis in hemimetabolous and holometabolous species. Scientific Reports. 1, 163 (2011).
- Philip, B. N., Tomoyasu, Y. Gene knockdown analysis by double-stranded RNA injection. Methods in Molecular Biology (Clifton, N. J). 772, 471-497 (2011).
- Zheng, Y., et al. CRISPR interference-based specific and efficient gene inactivation in the brain. Nature Neuroscience. 21 (3), 447-454 (2018).
- Garbutt, J. S., Bellés, X., Richards, E. H., Reynolds, S. E. Persistence of double-stranded RNA in insect hemolymph as a potential determiner of RNA interference success: Evidence from Manduca sexta and Blattella germanica. Journal of Insect Physiology. 59 (2), 171-178 (2013).
- Parrish, S., Fleenor, J., Xu, S., Mello, C., Fire, A. Functional anatomy of a dsRNA trigger: differential requirement for the two trigger strands in RNA interference. Molecular Cell. 6 (5), 1077-1087 (2000).
- Lemonds, T. R., Liu, J., Popadić, A. The contribution of the melanin pathway to overall body pigmentation during ontogenesis of Periplaneta americana. Insect Science. 23 (4), 513-519 (2016).
- Jackson, A. L., Linsley, P. S. Noise amidst the silence: Off-target effects of siRNAs. Trends in Genetics: TIG. 20 (11), 521-524 (2004).
- Patel, M., Peter, M. E. Identification of DISE-inducing shRNAs by monitoring cellular responses. Cell Cycle (Georgetown, Tex). 17 (4), 506-514 (2018).
- Ventós-Alfonso, A., Ylla, G., Montañes, J. C., Belles, X. DNMT1 promotes genome methylation and early embryo development in cockroaches. iScience. 23 (12), 101778 (2020).
- Wang, K., et al. Variation in RNAi efficacy among insect species is attributable to dsRNA degradation in vivo. Insect Biochemistry and Molecular Biology. 77, 1-9 (2016).
- Bi, F., Liu, N., Small Fan, D. interfering RNA: A new tool for gene therapy. Current Gene Therapy. 3 (5), 411-417 (2003).

