Detecting SARS-CoV-2 Virus by Reverse Transcription-Loop-Mediated Isothermal Amplification
Summary
Here we provide a complete protocol to standardize and implement the method for detecting the SARS-CoV-2 virus in human samples by reverse transcription loop-mediated isothermal amplification (RT-LAMP). This method, done within 60 min, could be adapted to any laboratory or point-of-care at a low cost and using inexpensive equipment.
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus has dramatically impacted human health. It continues to be a threat to modern society because many people die as a result of infection. The disease is diagnosed using serologic and molecular tests, such as the gold standard real-time polymerase chain reaction (RT-PCR). The last has several disadvantages because it requires specialized infrastructure, costly equipment, and trained personnel. Here, we present a protocol outlining the steps required to detect the SARS-CoV-2 virus using reverse transcription-loop-mediated isothermal amplification (RT-LAMP) in human samples. The protocol includes instructions for designing primers in silico, preparing reagents, amplification, and visualization. Once standardized, this method can be easily implemented and adapted to any laboratory or point-of-care within 60 min at a low cost and using inexpensive equipment. It is adaptable to detecting different pathogens. Thus, it can potentially be used in the field and in health centers to carry out timely epidemiological surveillance.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) causes coronavirus disease 2019 (COVID-19). The World Health Organization declared a public health emergency of international concern on 30 January 2020 and a pandemic on 11 March 2020. The pandemic resulted in over 760 million cases and 6.87 million deaths as of the date this article was written1.
The impact of this virus has highlighted the need for better, more accurate, faster, and more widely available surveillance tools to improve infectious disease detection and control2,3. During the pandemic, SARS-CoV-2 diagnostic tests were based on detecting nucleic acid, antibodies, and proteins, but RT-PCR detection of nucleic acid is the gold standard4. However, RT-PCR has some limitations; it requires specialized equipment, infrastructure, and personnel trained in molecular biology, limiting its application to specialized laboratories. Further, it is time-consuming (4-6 h), not including the time to transport the specimens to the laboratory, which can take days5. These constraints prevent efficient sample processing and obtaining the information required for contingency planning and epidemiological management.
Reverse transcription-loop-mediated isothermal amplification (RT-LAMP) has several advantages over RT-PCR, making it an appealing strategy for designing future point-of-care diagnostic tests (POCT), particularly in resource-constrained settings6. First, it is greatly specific because it uses between four and six primers that recognize six to eight areas in the target sequence, be it DNA or RNA7,8. Second, because it operates at a constant temperature, it does not require sophisticated equipment such as real-time thermal cyclers to generate the amplification, nor does it necessitate highly trained personnel to operate it. Third, the reaction time is very short (~60 min), and reagents that are not very specialized are employed, which makes it a cost-effective tool6. Given the foregoing and the health emergency caused by the COVID-19 pandemic, this technique can be viewed as an alternative diagnostic method that is quick, inexpensive, and simple to implement in any research laboratory9.
The protocol for standardizing and implementing an RT-LAMP to detect SARS-CoV-2 by colorimetric methods using a thermocycler and a water bath is described in this article (Figure 1). Critical points, their limitations, and alternatives to advance them are discussed.
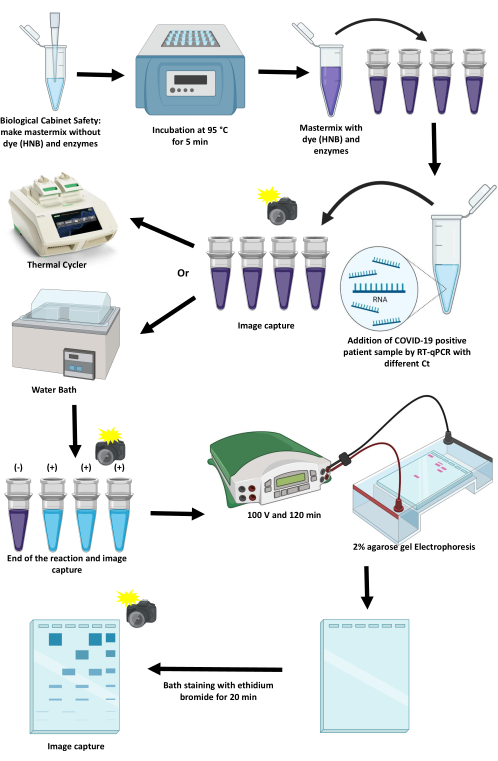
Figure 1: Scheme of the protocol for amplifying SARS-CoV-2 using the RT-LAMP technique. Please click here to view a larger version of this figure.
Protocol
Representative Results
Discussion
Although the RT-LAMP is regarded as a complementary methodology for performing molecular diagnostics, it also has some limitations and critical steps that must be considered when the protocol is standardized and implemented.
The LAMP standardization for the detection of SARS-CoV-2 evaluated the following parameters and components in the master mix: (a) concentration and temperature of alignment of the primers; (b) concentration of the enzymes; (c) magnesium concentration; (d) reaction time; (e…
Divulgazioni
The authors have nothing to disclose.
Acknowledgements
This work was funded by Sistema General de Regalías of Colombia, grant number BPIN 2020000100092, and Universidad Icesi – Convocatoria Interna, grant number CA0413119. MFVT was also financed by the Assistant Professorship Funds from Universidad de los Andes. The funding entities did not participate in the design, execution of activities, data collection, and data analysis and preparation of the manuscript. We thank to University Hospital Fundación Valle del Lili for viral RNA from Sars -CoV-2 samples and Dr. Alvaro Barrera-Ocampo for the comments on the manuscript.
Materials
| 1 kb DNA Ladder | SOLIS BIODYNE | 07-12-00050 | Store at -20 °C |
| 50x TAE Electrophoresis Buffer | ThermoScientific | B49 | Store at roome temperature |
| Accuris High Fidelity Polymerase | ACCURIS LIFE SCIENCE REAGENTS | PR1000-HF-200 | It can be used in case Q5 High-Fidelity DNA polymerase cannot be purchased. For the enzyme, make aliquots of an appropriate volume and store at -20 °C until use. |
| Agarose | PanReacAppliChem | A8963,0100 | N/A |
| Bst 3.0 DNA Polymerase 8000 IU/mL | New England BioLabs | M0374S/M0374L | For the enzyme, make aliquots of an appropriate volume and store at -20 °C until use. |
| Deoxynucleotide (dNTP) Solution Set | New England BioLabs | N0446S | Store at -20 °C |
| Diethyl pyrocarbonate | Sigma-Aldrich | 159220-25G | Handle it with caution under an extraction cabinet |
| GeneRuler 100 bp Plus DNA Ladder, ready-to-use | ThermoScientific | SM0322 | Store at -20 °C |
| Hydroxy naphthol blue disodium salt | Santa Cruz Biotechnology | sc-215156B | N/A |
| Q5 High-Fidelity DNA polymerase 2000 IU/mL | New England BioLabs | M0491S/M0491L | For the enzyme, make aliquots of an appropriate volume and store at -20 °C until use. |
| WarmStart RTx Reverse Transcriptase 15000 IU/mL | New England BioLabs | M0380S/M0380L | For the enzyme, make aliquots of an appropriate volume and store at -20 °C until use. |
Riferimenti
- World Health Organization. . Who coronavirus (COVID-19) dashboard (no date). , (2023).
- Ibrahim, N. K. Epidemiologic surveillance for controlling Covid-19 pandemic: types, challenges and implications. Journal of Infection and Public Health. 13 (11), 1630-1638 (2020).
- Rojas-Gallardo, D. M., et al. COVID-19 in Latin America: Contrasting phylodynamic inference with epidemiological surveillance. (Molecular epidemiology of COVID-19 in Latin America). medRxiv. , (2020).
- Liu, R., et al. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clinica Chimica Acta. 505, 172-175 (2020).
- Kevadiya, B. D., et al. Diagnostics for SARS-CoV-2 infections. Nature Materials. 20 (5), 593-605 (2021).
- Tomita, N., Mori, Y., Kanda, H., Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nature Protocols. 3 (5), 877-882 (2008).
- Li, Y., Fan, P., Zhou, S., Zhang, L. Loop-mediated isothermal amplification (LAMP): A novel rapid detection platform for pathogens. Microbial Pathogenesis. 107, 54-61 (2017).
- Notomi, T., Mori, Y., Tomita, N., Kanda, H. Loop-mediated isothermal amplification (LAMP): principle, features, and future prospects. Journal of Microbiology. 53 (1), 1-5 (2015).
- Augustine, R., et al. Loop-mediated isothermal amplification (LAMP): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology (Basel). 9 (8), 182 (2020).
- . Nextstrain Available from: https://nextstrain.org/ (2023)
- . Neb Lamp, NEB LAMP Available from: https://lamp.neb.com/ (2023)
- . Blast: Basic local alignment search tool (no date) Available from: https://blast.ncbi.nlm.nih.gov/ (2023)
- Zhang, Y., et al. Rapid molecular detection of SARS-CoV-2 (COVID-19) virus RNA using colorimetric LAMP. medRxiv. , (2020).
- Lu, R., et al. Development of a novel reverse transcription loop-mediated isothermal amplification method for rapid detection of SARS-CoV-2. Virologica Sinica. 35 (3), 344-347 (2020).
- Najafov, A., Hoxhaj, G. . PCR Guru. , (2017).
- Zhang, Y., et al. Enhancing colorimetric loop-mediated isothermal amplification speed and sensitivity with guanidine chloride. Biotechniques. 69 (3), 178-185 (2020).
- Ramírez-Chavarría, R. G., et al. Automatic analysis of isothermal amplification via impedance time-constant-domain spectroscopy: A SARS-CoV-2 case study. Chemosensors. 11 (4), 230 (2023).
- Haque, M. F. U., et al. A novel RdRp-based colorimetric RT-LAMP assay for rapid and sensitive detection of SARS-CoV-2 in clinical and sewage samples from Pakistan. Virus Research. 302, 198484 (2021).
- Donia, A., et al. Integration of RT-LAMP and microfluidic technology for detection of SARS-CoV-2 in wastewater as an advanced point-of-care platform. Food and Environmental Virology. 14, 364-373 (2022).

