Quantification of Colonic Stem Cell Mutations
Summary
We report significant improvements for the reproducible measurement of somatic colonic stem cell mutations after exposure of mice to potential DNA damaging agents.
Abstract
The ability to measure stem cell mutations is a powerful tool to quantify in a critical cell population if, and to what extent, a chemical can induce mutations that potentially lead to cancer. The use of an enzymatic assay to quantify stem cell mutations in the X-linked glucose-6-phosphate dehydrogenase gene has been previously reported.1 This method requires the preparation of frozen sections and incubation of the sectioned tissue with a reaction mixture that yields a blue color if the cells produce functional glucose-6-phosphate dehydrogenase (G6PD) enzyme. If not, the cells appear whitish. We have modified the reaction mixture using Optimal Cutting Temperature Compound (OCT) medium in place of polyvinyl alcohol. This facilitates pH measurement, increases solubilization of the G6PD staining components and restricts diffusion of the G6PD enzyme. To demonstrate that a mutation occurred in a stem cell, the entire crypt must lack G6PD enzymatic activity. Only if a stem cell harbors a phenotypic G6PD mutation will all of the progeny in the crypt lack G6PD enzymatic activity. To identify crypts with a stem cell mutation, four consecutive adjacent frozen sections (a level) were cut at 7 µm thicknesses. This approach of making adjacent cuts provides conformation that a crypt was fully mutated since the same mutated crypt will be observed in adjacent sections. Slides with tissue samples that were more than 50 µm apart were prepared to assess a total of >104 crypts per mouse. The mutation frequency is the number of observed mutated (white) crypts ÷ by the number of wild type (blue) crypts in a treatment group.
Introduction
Colon cancer is thought to involve exposure to environmental agents and dietary components that can damage DNA and produce activating somatic mutations in oncogenes (e.g., ß-catenin) or inactivating mutations in tumor suppressor genes (e.g., APC). It is assumed that these critical mutations occur in colonic stem cells. Because of the unique crypt architecture of the colonic epithelium, it is possible to measure stem cell mutations in the colon after exposing animals to chemicals associated with colon carcinogenesis. Several X-linked enzymes can serve as indicators for mutations that occur in stem cells, as the mutations will be present in all cells within a crypt.
Previously, a procedure was published demonstrating that chemicals that induce colon tumors in mice also generated somatic stem cell mutations in the colon via analysis of mutations in the X-linked glucose-6-phosphate dehydrogenase (G6PD) gene.1-3 The method quantifies the incidence of random somatic mutations in colonic stem cells without any selection pressure. The procedure involves the production of unfixed frozen colon sections from treated and control mice and the identification of crypts devoid of cells that produce functional G6PD activity. These mutated crypts, which appear white, indicate that the mutation occurred in a stem cell that gave rise to progeny also harboring a mutated G6PD gene. In the enzymatic assay, G6PD deficient mutant crypts cannot oxidize glucose-6-phosphate, which is required for the reduction of nitro blue tetrazolium (NBT). When the G6PD enzyme is functional, the NBT in the staining mixture is reduced to insoluble formazan and precipitates, accumulating at the location of the enzyme and “staining” cells blue. Cells that are G6PD mutants remain whitish in contrast to blue stained wild type crypts. This method measures mutations that have a “null” phenotype. Because enzymes, such as G6PD, can diffuse out of the cells on an unfixed tissue section if the sections are placed in an aqueous solution, it is necessary to stabilize the enzyme in the tissue sections to be analyzed.4 The stabilization of the enzyme in the tissue must not interfere with diffusion of small reagent molecules necessary for the G6PD enzymatic reaction.
We have made a number of significant changes to the original procedure. The medium for the critical enzymatic staining has been changed from polyvinyl alcohol to Optimal Cutting Temperature Compound (OCT), which facilitates pH control and solubilization of the components used in the assay. The staining is performed using 0.4 – 0.5 ml wells made of steel washers so that each tissue section received the same volume of the G6PD reaction mixture. A procedure was developed to estimate the number of crypts in an imaged section that provides a large sampling of the colonic tissue without having to manually count >105 crypts for each colon. The analysis of adjacent 7 µm tissue sections affords visualization of a mutant crypt on more than one slide, which reduces potential staining artifacts. These changes make the procedure more efficient and reproducible.
Using this revised protocol, we have quantified the mutation frequency in colonic stem cells after exposure of C57Bl/6 mice to azoxymethane, a well-known colon carcinogen in rodents.5-7
Protocol
The experimental procedures and ethical treatment of animals was approved by the University of Pittsburgh IACUC (protocol #1104674).
1. Preparation of G6PD Staining Mixture
NOTE: Ensure that the final concentration of each reagents is as follows; 5 mM glucose-6-phosphate (G6P); 2 mM NADP, 5 mM MgCl2, 0.35 mM 1-methoxy-5-methylphenazinium methyl sulfate (MMPMS) and 0.8 mM NBT.1,8,9 For each reagent the initial concentration was derived for a 40 ml final volume. Solutions were freshly prepared and used each day.
- Add 2 ml of 200 mM pH 7.4 phosphate buffer (PB) to 35 ml of OCT medium in a 50 ml tube and shake vigorously. Confirm with pH paper that the solution is ~pH 7.4. If not discard the solution.
- Add 61 mg G6P dissolved in 0.94 ml of 200 mM PB, 61 mg NADP dissolved in 0.94 ml of 200 mM PB, and 41 mg MgCl2 dissolved in 0.96 ml of 100 mM PB. Reconfirm that the pH is ~7.4 with pH paper. If not ~7.4, discard the solution.
- Add 5 mg of MMPMS dissolved in 0.25 ml of 200 mM PB (pH 7.4). Vigorously shake the resulting reaction mixture to generate a clear dark red homogenate. If the solution is turbid, the mixture should be discarded. The tube containing the reaction mixture is wrapped in foil to minimize exposure to light. Store the reaction mixture at RT while the final component, NBT, is prepared.
- Prepare separately a 5 mM NBT solution by adding 0.4 ml anhydrous dimethyl formamide (DMF) and 0.4 ml anhydrous ethanol to 164 mg of NBT in a 2 ml tube with an O-ring lid. Close the lid loosely (not tightly sealed) and bring the solution to a vigorous boil in an oil bath until the NBT is in solution.8
- Allow the solubilized NBT to cool to RT and then add all of the solution to the OCT reaction mixture and shake vigorously to obtain a clear solution. Observe the solution color change from initial orange-red to a dark-red or purple color over time. This is normal.
- Place the final G6PD staining mixture in a 37 °C warm room for 1 hr before use.
2. Animal Treatment and Tissue Collection
- Treat 6-week-old C57Bl/6 male mice with DNA damaging agent, e.g., 10 mg/kg azoxymethane (AOM) in a volume of 200 µl PBS by i.p. injection. Treat control mice with 200 µl of PBS by i.p. injection.
- At 90 days, euthanize mice by CO2 asphyxiation followed by cervical dislocation as approved by IACUC protocol.
- Surgically open the abdominal cavity from just above the anus to the base of the sternum. Move the small intestine out of the body cavity but do not excise it. Excise the lower intestine just below the cecum to the rectum.10
- Carefully slice along one side of the colon using a micro-dissection scissor and remove feces from the opened colon by washing with clean sterile PBS. Swiss roll the excised colon with the luminal side facing outward, and place in a tissue mold.10
- Completely immerse the colon in OCT medium and place mold in a Dewar containing liquid N2. Hold the mold so that the plastic base is just in contact with the liquid N2 until the OCT medium is frozen solid.
- Store the frozen tissue block at -80 until cutting frozen sections. Do not use any fixative, which would result in the inactivation of the G6PD enzyme.
3. Preparation of Frozen Sections
- Cut frozen tissue sections at a thickness of 7 µm using a cryostat at -25 °C. Cut 4 consecutive 7 µm sections and place two of them on the same slide (Figure 1). Prepare 2 slides, or four consecutive 7 µm sections to represent one level of the colon (Figure 1).
- Repeat for the next level with a minimum distance of >50 µm from the last section of the previous level. This distance ensured that the crypts assessed for each level did not duplicate one another.
- Frequently wipe the cryostat blade with 100% ethanol to remove the OCT residue that accumulates during cutting. Before sectioning is resumed, the dry blade is wiped with an antistatic dryer sheet to dissipate accumulated static charge. Store slides with the frozen sections at -80 oC.
4. Staining of Frozen Section Tissue
- Perform the enzyme histochemistry reaction in a 37 °C warm room with high humidity.
NOTE: Attempting the enzymatic reaction in an incubation oven or on a slide warmer led to poor and inconsistent G6PD staining. - Build wells for tissue staining using two 1.5 cm x 0.15 cm steel washers (interior diameter x height) that are bonded together using silicone grease (Figure 1). Cut the parafilm to fit the base of the well with the center cut out and the well placed over the tissue section on the slide.
NOTE: The parafilm on the bottom of the washer creates a seal between the slide and the washer that maintains the viscous G6PD staining mixture on the tissue section. This construct gives a well with a 0.4 – 0.5 ml volume so that each tissue section in the study receives the same volume of the G6PD reaction staining mixture. - Incubate frozen tissue slides at 37 oC in a warm room for 10 min. At 37 °C, add the G6PD staining reaction mixture to each tissue section well (2 sections per slide) for 45-50 min. Remove slides from warm room, and carefully lift the stainless steel wells off the slides.
- Set slides on their long edge to allow the reaction medium to drain. Place slides in 100 mM PB (pH 7.4) for 30-60 min to remove the G6PD staining reaction mixture from the tissue without disturbing the tissue staining. Submerge slides in distilled water for 5-10 min to remove PB salts
- Seal the tissue by adding fluoro-gel from a dropper onto the tissue and waiting ~ 30 min for it to dry. Avoid bubbles that can confound the identification of mutated crypts.
NOTE: If bubbles form, the procedure can be repeated after removal of the fluoro-gel by soaking in distilled water. Do not use cover slips as they tend to trap air bubbles.
5. Determination of G6PD Mutant Crypts and Mutation Frequency
- Analyze slides for G6PD mutant crypts under a light microscope.
NOTE: The criteria used to identify a fully mutant crypt were: (i) the entire crypt was devoid of blue color; (ii) the outer structure of the crypt was intact, (iii) the mutant crypt was observed on adjacent 7 µm sections and, (iv) two observers had to identify the same crypt as a mutant. Mutants observed on adjacent 7 µm sections are counted as one mutant. - Image each G6PD mutant crypt at 40x magnification and catalogue the images using a 5.0 megapixel camera with imaging software.
- Quantify the total number of crypts examined in a mouse by selecting a representative section for each mouse from the level with the smallest surface area and image it at 10x magnification (Figure 1C).
NOTE: A level represents four consecutive 7 µm sections (Figure 1D), which due to their proximity often show mutated crypts on more than one section. The levels are separated by >50 µm to visualize crypts from different areas of the colon. - Open each image file and visually count the number of crypts on a level by analyzing the files (shown as boxes in Figure 1C). Images files will vary in the number of crypts observed from as low as 20 to >300 crypts in one file.
- Multiply the total number of crypts from the selected level (Figure 1C) for each mouse by the number of levels screened for G6PD mutant crypts for each colon. For example, a total count of 1,250 crypts per selected level multiplied by 8 levels screened for mutant crypts equals a total count of 10,000 crypts. Evaluate a minimum of 10,000 crypts for each colon for statistical analysis.
- Combine the number of mutants observed in all of the animals by the total number of crypts screened to calculate the MF for each treatment. The background stem cell MF level in untreated mice is < 0.1×10-4 (Table 1), which is close to that previously reported in C57Bl/6 mice.11
Representative Results
The ability to measure colonic stem cell mutations in animals provides a unique way to correlate mutations to cancer induction. Normally, it is assumed that the critical step in carcinogenesis involves activating mutations in oncogenes and/or inactivating mutations in tumor suppressor genes. We injected C57Bl/6 mice with 200 µl of PBS or 10 mg/kg AOM in 200 µl of PBS. AOM is a known colon carcinogen.5-7 At 90 days the colons were analyzed for stem cell mutations. This time is required to allow stem cells to fully populate a crypt, and only when an entire crypt does not produce G6PD is a crypt identified as a stem cell mutant. Examples of fully mutant crypts are shown in Figure 2. The figure shows two different orientations (vertical and horizontal) of the crypts that occur in the preparation of the frozen sections. The vertical view is looking down the long axis of the crypt while the horizontal provides a side view. The crypts that are wild type for G6PD stain blue with some white that is due to mucous production from goblet cells. By counting the total number of crypts on a slide (Figure 1) and the number of mutated crypts, the MF for mutated crypts can be calculated. No mutated crypts were observed in any of the >100,000 wild type crypts inspected in the control (PBS treated) mice. We set the MF for the control mice to be < 0.1×10-4, which is close to that reported previously for C57Bl/6 mice.1
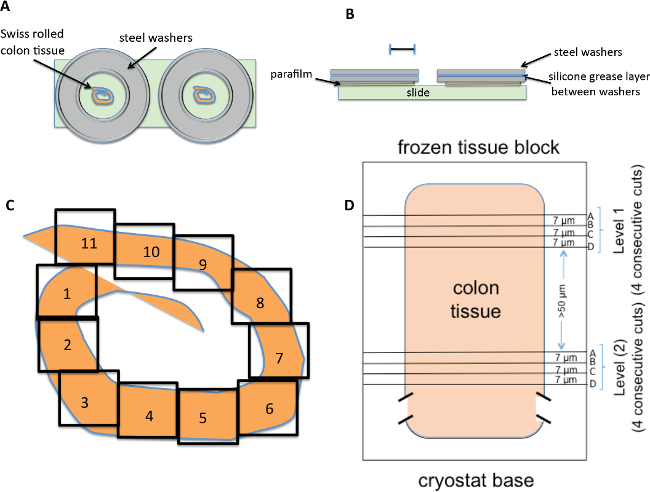
Figure 1. Preparation of slides for G6PD staining reagent. (A) Two 1.5 cm x 0.15 cm steel washers (interior diameter x height) are sealed together using silicone grease. Parafilm is then cut to fit the base of the well with the center cut out and the well placed over the Swiss rolled colon tissue sections on the slide. (B) Two tissue sections from adjacent 7 µm cuts are placed on the same slide and stained at the same time. Scale bar: 1.0 cm. (C) The number of crypts in the section is determined by looking at the tissue at 10x magnification. The number of fields (shown as boxes) varies with each section. The number of crypts in each field is determined by visual counting and added to give the total number of crypts for that level. (D) A level is defined as four consecutive 7 µm tissue sections. The levels are > 50 µm apart to avoid overlap with previously assessed crypts. Please click here to view a larger version of this figure.
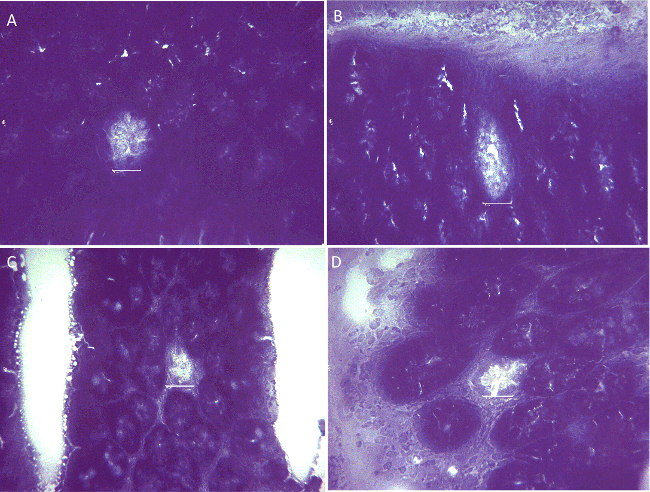
Figure 2. Representative examples of mutated crypts observed in AOM treated C57Bl/6 male mice that do not produce functional G6PD. The G6PD mutant crypts are well defined but are virtually devoid of blue stain. Wild type crypts that comprise most of the crypts in the field are blue/purple indicating G6PD enzymatic activity. Scale: 10 µm. Please click here to view a larger version of this figure.
| treatment | incidence of mutant crypts | mutant cryptsa | total crypts analyzed | M.F. (x 10-4) |
| solvent | 0/6 mice | 0 | > 105 | < 0.10 |
| AOM | 10/12 mice | 43 | 96821 | 4.44 |
Table 1. Mutation frequency in untreated and azoxymethane treated C57Bl/6 mice.12
Colonic stem cell mutations in WT C57Bl/6 mice 90 days after exposure to 10 mg/kg azoxymethane (AOM) or solvent control.
Discussion
Often the genotoxic effect of a compound is determined by its ability to modify DNA. This is normally done by isolating the tissue and measuring the global level of DNA adducts. For AOM, this would involve quantifying O6-methylguanine adducts in the colon. Using this approach information on damage within specific cell types, such as in the stem cell niche, is lost. In addition, a DNA adduct is not the same as a mutation since only a small subset of adducts are eventually converted into fixed mutations.12 We provide herein the details to reproducibly quantify stem cell mutations in the colon based on the initial method described by Griffiths et al.1 and based on G6PD staining developed by VanNorden.4,8,9,13,14
There are several critical steps for this enzyme histochemistry method to produce a quantifiable result. One is in the sectioning of the tissues. Significantly more tissue sections must be generated and analyzed than what would be typically done with H&E histological analysis. We determined that 7 µm sections afforded the best G6PD staining after evaluation of 5, 6, 7, 8 and 10 µm thick sections.
Secondly, staining the necessary number of valuable tissue sections required a reproducible reaction mixture. We found the use of polyvinyl alcohol (PVA) used in the original protocol to be problematic because it is quite viscous at the solution percentages (18% and greater) necessary to facilitate the enzyme histochemistry reaction while preventing the diffusion of the G6PD enzyme from the tissue. Better solubilization of the reagents and control of the pH was accomplished by replacing the PVA with OCT medium, which contains 10% PVA. The pH could be accurately determined with pH paper and if the pH is not close to 7.4, the solution is discarded without wasting precious tissue sample and time. This change led to better and more consistent staining in the tissue sections. At the pH (7.2-7.4) required to measure G6PD activity, the complete reaction mixture has a characteristic clear deep orange-red color, which will slowly change to purple. Regardless of the color, if the reaction staining solution is turbid it should be discarded since it will not afford good staining. We found that some lots of OCT medium provided mixtures outside the required pH range so it is advisable to test lots by adding the PB (7.4) to the OCT medium and determining the pH. We also used MMPMS as previously suggested,8,9 rather than the 5-methylphenazenium methyl sulfate used in the original paper1 to eliminate the light sensitivity of the G6PD staining reaction mixture.
Finally, the method for deriving the MF is novel compared to other methods previously employed.1,2 Staining artifacts can lead to the potential identification of false G6PD mutant crypts. To troubleshoot this problem, we analyzed 4 contiguous 7 µm tissue sections that allows for visualization of the same mutated crypt in more than one tissue section (Figure 4D). This verification reduces the chances of staining artifacts affecting the MF analysis. Also, our method uses an actual value of total crypts counted to estimate the total number of crypts analyzed when calculating the MF.
Because of the unique crypt structure found in the small and large intestine, it is possible to determine the location of stem cells and cell lineage for each crypt. A major limitation to the approach is that it is restricted to the intestines because the morphology of other tissues is not as well defined as in the colon. Therefore, it is not possible to determine the stem cell MF in most other tissues that are susceptible to cancer. Since the assay is a phenotypic assay based on the loss of G6PD enzymatic activity, the actual MF in the stem cell population will be higher than we report for G6PD due to mutations at wobble bases and point mutations that do not significantly affect enzymatic activity.
While there are limitations and challenges to staining for G6PD activity in colon tissue to quantify somatic stem cell mutations, significant modifications were introduced in this paper to enhance the feasibility of utilizing this method. We found that these changes enhanced the reproducibility of generating the G6PD staining reagent and quantifying the MF. Moreover, staining for G6PD enzyme activity as a marker for somatic stem cell mutations does not require an expertise in immunohistochemistry staining as would testing for the expression of metallothionein,15 another potential marker of somatic stem cell mutations.
In the future, we will analyze colonic stem cell mutations in mice treated with environmental compounds, such as 2-amino-1-methyl-6-phenylimidazo[4,5-b]pyridine (PhIP) and other aromatic amines found in grilled meats, that are associated with human colon cancer.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors have no acknowledgements.
Materials
| reagents | |||
| nitroblue tetrazolium | |||
| NADP | |||
| glucose-6-phosphate | |||
| 1-methoxy-5-methylphenazinium methyl sulfate | |||
| dimethyl formamide | |||
| phosphate buffer (pH 7.4 | |||
| Optimal Cutting Temperature Compound (OCT) | |||
| Equipment | |||
| light microscope equipped with 5 megapixel camera | |||
| cryostat | |||
| warm room |
References
- Griffiths, D. F., Davies, S. J., Williams, S., Williams, G. T., Williams, E. D. Demonstration of somatic mutation and colonic crypt clonality by X-linked enzyme histochemistry. Nature. 333 (6172), 461-463 (1988).
- Griffiths, D. F., Sacco, P., Williams, G. T., Williams, E. D. The clonal origin of experimental large bowel tumours. Br. J. Cancer. 59 (3), 385-387 (1989).
- Kuraguchi, M., Thomas, G. A., Williams, E. D. Somatic mutation of the glucose-6-phosphate dehydrogenase (g6pd) gene in colonic stem cells and crypt restricted loss of G6PD activity. Mutat. Res. 379 (1), (1997).
- Van Noorden, C. J., Vogels, I. M. Polyvinyl alcohol and other tissue protectants in enzyme histochemistry: a consumer’s guide. Histochem. J. 21 (7), 373-379 (1989).
- Giardina Rosenberg, D. W., C, T., Tanaka, Mouse models for the study of colon carcinogenesis. Carcinogenesis. 30 (2), 183-196 (2009).
- Tanaka, T., Kohno, H., Suzuki, R., Yamada, Y., Sugie, S., Mori, H. A novel inflammation-related mouse colon carcinogenesis model induced by azoxymethane and dextran sodium sulfate. Cancer Sci. 94 (11), 965-973 (2003).
- Suzuki, R., Kohno, H., Sugie, S., Tanaka, T. Dose-dependent promoting effect of dextran sodium sulfate on mouse colon carcinogenesis initiated with azoxymethane. Histol. Histopathol. 20 (2), 483-492 (2005).
- Frederiks, W. M., Vreeling-Sindalarova, H., Van Noorden, C. J. Loss of peroxisomes causes oxygen insensitivity of the histochemical assay of glucose-6-phosphate dehydrogenase activity to detect cancer cells. J. Histochem. Cytochem. 55 (2), 175-181 (2007).
- Hisada, R., Yagi, T. 1-Methoxy-5-methylphenazinium methyl sulfate. Biochem. 82 (5), 1469-1473 (1977).
- Whittem, C. G., Williams, A. D., Williams, C. S. Murine Colitis modeling using Dextran Sulfate Sodium (DSS). J Vis Exp. (35), (2010).
- Kuraguchi, M., Cook, H., Williams, E. D., Thomas GA, . Differences in susceptibility to colonic stem cell somatic mutation in three strains of mice. J. Pathol. 193 (4), 517-521 (2001).
- Whetstone, R. D., Gold, B. T-Cells Enhance Stem Cell Mutagenesis in the Mouse Colon. Mutat. Res. 744, 1-5 (2015).
- Van Noorden, C. J., Vogel, I. M. Histochemistry and cytochemistry of glucose-6-phosphate dehydrogenase. Prog. Histochem. Cytochem. 15 (4), 1-85 (1985).
- Van Noorden, C. J., Vogels, I. M. A sensitive cytochemical staining method for glucose-6-phosphate dehydrogenase activity in individual erythrocytes. II. Further improvements of the staining procedure and some observations with glucose-6-phosphate dehydrogenase deficiency. Br. J. Haematol. 60 (1), 57-63 (1985).
- Cook, H. A., Williams, D., Thomas, G. A. Crypt-restricted metallothionein immunopositivity in murine colon: validation of a model for studies of somatic stem cell mutation. J. Pathol. 191 (3), 306-312 (2000).

