Air-sampled Filter Analysis for Endotoxins and DNA Content
Summary
Two complementary analyses of atmospheric biological particles from air sampled filters are described herein: the extraction and detection of endotoxin, and of DNA.
Abstract
Outdoor aerosol research commonly uses particulate matter sampled on filters. This procedure enables various characterizations of the collected particles to be performed in parallel. The purpose of the method presented here is to obtain a highly accurate and reliable analysis of the endotoxin and DNA content of bio-aerosols extracted from filters. The extraction of high molecular weight organic molecules, such as lipopolysaccharides, from sampled filters involves shaking the sample in a pyrogen-free water-based medium. The subsequent analysis is based on an enzymatic reaction that can be detected using a turbidimetric measurement. As a result of the high organic content on the sampled filters, the extraction of DNA from the samples is performed using a commercial DNA extraction kit that was originally designed for soils and modified to improve the DNA yield. The detection and quantification of specific microbial species using quantitative polymerase chain reaction (q-PCR) analysis are described and compared with other available methods.
Introduction
Air sampling on filters is a basic tool in atmospheric aerosols research.1 The sampled filters are the starting point for various chemical, physical, and biological characterizations of the collected ambient particles.2-11 The advantage of this approach is that various analyses can be performed off-line on the same sample. Compiling the data from all the different analyses enables the researcher to obtain a good understanding of the characteristics of the collected particles and aids in solving complex questions in the atmospheric sciences.12,13 For example, marine and inland air-samples taken during the same period can be compared with respect to the sampled particle toxicity and biological composition.14 For this study, lipopolysaccharides (LPS), components on gram-negative bacterial cell-walls, also known as endotoxins, were extracted from filters sampled on-shore and at an inland site, and were evaluated using the limulus amebocyte lysate (LAL) test. In parallel, a genomic evaluation of the bacterial content (total bacteria, gram negative, and cyanobacteria) was performed on the same sample using the quantitative polymerase chain reaction (q-PCR). The LAL test is based on measurements of turbidity formed following the addition of an aqueous extract of amebocytes from the horseshoe crab, Limulus polyphemus, to an aqueous solution containing the endotoxins. The higher the endotoxin concentration in the sample, the faster turbidity develops.15 The q-PCR analysis is based on a fluorescence signal emitted as a specific DNA fragment is amplified.16 By real time monitoring of the signal during the exponential phase of the PCR reaction and calibrating with a standard curve, the initial DNA amount can be quantified. The combination of these two analyses together with others, as detailed elsewhere,14 can provide a good estimation of the levels of endotoxin and the amount of the source bacteria in the samples.
The purpose of the method presented here is to obtain a highly accurate and reliable analysis of the endotoxin and DNA content of bio-aerosols extracted from filters. While methods for sampling the physical and inorganic chemical characteristics of aerosols are well known and, more recently, methods have been developed to investigate its organic matter component,17 there has been scant research on the biological component of aerosols.18 The rationale for the current method is to address this gap by presenting in detail a robust method for extracting, analyzing, and identifying the biological fraction of airborne aerosols.14
The method detailed here is expected to find wide-spread use in biological indoor and outdoor aerosol research projects involving filter analysis.20-24
Protocol
Note: A detailed list of all the materials and instruments used in this protocol is shown in the Materials section.
1. Air Sampling on Filters
- Preparation of Filters
- For high volume sampling, use 20.3 x 25.4 cm2 filters. Choose the specific type of filter that best fits the research needs, as well as the filter cut-off size, if applicable.1 Here, use quartz microfiber filters.
- Pre-bake filters intended for organic and biological compound sampling to destroy organic residues. To pre-bake the filters, wrap them individually in aluminum foil and then bake them in a laboratory furnace at 450 °C for at least 5 hr.
- Store the baked filters at -20 °C until sampling.
- Instrument Handling and Air Sampling
- Clean residual dust from the head of the high volume sampler. Wipe the parts with clean paper wipes and allow them to air-dry before reconnecting them to the sampler.
- Disinfect the filter cassette with ethanol, and work with gloves and a lab coat at all times.
- Place a pre-baked filter inside the clean filter cassette, and proceed according to the high volume sampler manual.
- Mark the date and any unusual weather conditions, if applicable (e.g. rain, dust storm) on the aluminum wrap, and save it in a clean place until the completion of sampling.
- Collect the sampled filter and fold it in half, with the sampled side facing inward.
- Wrap the filter in the original aluminum foil and store at -20 °C until analysis.
Note: If the time lag between sampling and analysis is longer than 2 months, store the sample at -80 °C to avoid degradation of the organic and biological material.
2. Endotoxin Analysis
Note: Disinfect the work surface with 70% ethanol and work with pyrogen-free tubes, tips and reagents only. If glassware are used, pre-heating at 250 °C for 30 min, or 200 °C for 60 min is required.25 Prepare all reagents in a class II biosafety cabinet and work with gloves and a lab coat at all times.
- Reagent Preparation
- Follow the LAL kit manufacturer's protocol to rehydrate the lysate shortly before use.26 Tap gently on the vial to dislodge LAL remaining on the bottle walls. Lift the stopper gently to break the vacuum. Using a needled syringe, add 5 ml of pyrogen free water (PFW) to the vial to rehydrate all the lysate content and mix gently to avoid foam formation.
- Seal the vial with plastic paraffin film when not in use and store at 4 °C for up to two days. Store any remaining lysate at -20 °C for up to three months.
Note: This reagent can be frozen only once. - Rehydrate the control standard endotoxin (CSE), which contains 0.5 µg E. coli, in accordance with the manufacturer's protocol.27 Determine the specific amount of water to be added to the CSE vial from the manufacturer's website28 by entering the lot number for the kit where specified on the website. The final concentration of E. coli in the vial, is measured in endotoxin units (EU; where 10 EU = 1 ng). Label this vial Ed0.
- Seal the CSE vial with plastic paraffin film when not in use and store at 4 °C for up to 4 weeks.
- Standard Curve and Spiked-filter Preparation
- In a class II biosafety cabinet, prepare 22 pyrogen-free 2 ml centrifuge tubes containing decreasing amounts of CSE (tubes Ed1-Ed11) or no CSE (the blank tube containing only PFW) to produce a duplicated endotoxin dilution series, as shown in Table 1.
- In a biological cabinet with circular flow, cut 22 circular pieces of clean, pre-baked filter using a disinfected 1.12 cm diameter cork borer.
- Place the filter pairs into sterile petri dishes.
- Spike 50 µl from each member of endotoxin dilution series Ed2-Ed11 and from the blank tube onto a corresponding pair of filters.
- Leave the Petri dishes containing the spiked filters open under the hood to dry for 30 min.
- Place each filter piece in a pyrogen-free 2 ml tube.
| Tube | Final endotoxin concentration (EU ml-1) | Standard endotoxin volume (ml) | Pyrogen free water volume (ml) | total volume (ml) |
| Ed0 | 2,500 | stock solution* | 5 | |
| Ed1 | 1,000 | 80 (from Ed0) | 120 | 200 |
| Ed2 | 100 | 20 (from Ed1) | 180 | 200 |
| Ed3 | 50 | 10 (from Ed1) | 190 | 200 |
| Ed4 | 25 | 5 (from Ed1) | 195 | 200 |
| Ed5 | 12.5 | 2.5 (from Ed1) | 197.5 | 200 |
| Ed6 | 6.25 | 1.25 (from Ed1) | 198.75 | 200 |
| Ed7 | 3.125 | 6.25 (from Ed2) | 193.75 | 200 |
| Ed8 | 1.563 | 6.25 (from Ed3) | 193.75 | 200 |
| Ed9 | 0.781 | 6.25 (from Ed4) | 193.75 | 200 |
| Ed10 | 0.391 | 6.25 (from Ed5) | 193.75 | 200 |
| Ed11 | 0.195 | 6.25 (from Ed6) | 193.75 | 200 |
| Blank | 0 | 0 | 200 | 200 |
| * Prepare the stock solution as per the manufacturer's instructions. | ||||
Table 1: Endotoxin Standard Curve. Endotoxin concentration, volume of standard endotoxin and of pyrogen-free water to be added, and the total volume obtained are detailed for each dilution tube in the calibration curve.
- Endotoxin Extraction from the Experimental and Spiked Filters
- In a biological cabinet with circular flow, cut the sample filters (from Step 1.2) into circular pieces using a disinfected 1.12 cm diameter cork borer, and place them, together with the spiked standard filters (from Step 2.2), in pyrogen-free 2 ml tubes (i.e., the sample tubes).
- Add 1 ml PFW to the tubes and shake them for 60 min at room temperature, using a laboratory shaker.
- Centrifuge the sample tubes in a microcentrifuge at 375 x g for 10 min.29
- Transfer the supernatant, which contains the endotoxins, into a new pyrogen-free 2 ml tube.
- Limulus Amebocyte Lysate (LAL) Test
- Determine the endotoxin concentration in the samples by performing the LAL test in a pyrogen-free 96-well microplate with a flat bottom and a lid.
- Plan the plate array in advance, as shown in Figure 1. Include a standard curve prepared directly from endotoxin standard solutions Ed2-Ed11 (and thus covering the concentration range of 100-0.195 EU/ml (see section 2.2)) in every running plate. The standard curve should occupy wells 1-10 of rows A and B of the plate. Set aside wells 11-12 in these rows for blank samples (making a total of four blanks).
- Turn on the microplate reader and program it for a kinetic reaction involving incubation of the microplate at 37 °C and shaking every 5 min, followed by absorption measurements at 405 nm. Repeat 18 times for 1.5 hr.
- Determine the efficiency with which the endotoxins are extracted from the sample filters (i.e., the samples obtained from the spiked standard filters) via division of the calculated endotoxin amount by the original amount spiked.
- Start the assay by placing 50 µl of the standard endotoxin solutions (from step 2.4.2), or of the spiked filter extract and its blanks (from section 2.3), or of the experimental samples and its blanks (also from section 2.3), as per the planned plate-array (Figure 1) and close the cover.
- To each well, quickly add 50 µl of LAL solution, and gently shake the plate horizontally while it is placed on the table before lifting it into the reader.
- Place the plate in the plate reader and start the experimental run.
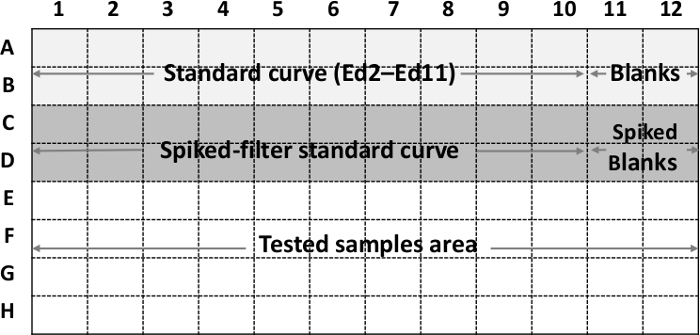
Figure 1: Endotoxin array plate. An example of an endotoxin analysis array in a 96-well microplate.
3. Genomic Analysis
Note: For DNA extraction, disinfect the work surface with 70% ethanol and work with sterile tubes, tips and reagents only. For q-PCR analysis of DNA, disinfect the work surface with surface DNA-decontaminant. Prepare all reagents in a class II biosafety cabinet and work with gloves and a lab coat at all times.
- Preparation of the DNA Primers and Probes
- Prior to DNA extraction, either order a commercially-available set of primers and a probe (if the hydrolysis probe method is applied), or design a new set of primers and a probe using primer designing computational tools.30
- Rehydrate the primers according to the manufacturer's protocol using either 10 mM Tris-0.1 mM Ethylenediaminetetraacetic acid (EDTA) (TE buffer) or PCR-grade water.
Note: It is recommended to dissolve the initial PCR primer stock in TE buffer with low EDTA (0.1%) as it prevents primer degradation when kept for longer times. Use PCR-grade water for subsequent dilutions to reduce EDTA amounts that might inhibit PCR reactions. - Prepare aliquots of 50 µl or 100 µl of the primer and probe in sterile 0.5 ml tubes and store at -20 °C until analysis.
- Standard Cell Concentration Evaluation Prior to DNA Extraction
- In a suitably sized centrifuge tube, prepare a standard cell solution of the microbial species of interest (tube Od0, Table 2A). Mix the cell suspension by pipetting it up and down in the tube 7-10 times using a pipette with a small bore.
- If the cells are colored (e.g. certain fungal spores), do not perform a staining procedure. If cell staining is required, dilute the cell suspension in a suitable dye for microscopic detection of the cells of interest.31
- Clean the cover slip and hemocytometer with ethanol. Moisten and affix the cover slip to the hemocytometer.
- Load about 8 µl of the cell suspension into both hemocytometer chambers by carefully touching the edge of the cover slip with the tip of the pipette and filling the chamber by capillary action. Do not over/under fill the chamber.
- Determine the number of cells by viewing them under a microscope at 400X magnification (40X in objective and 10X in ocular). Nine squares measuring 1 x 1 mm2 and arranged in a 3 x 3 grid should be seen. Focus the microscope on one of the four corner squares of the grid (a higher magnification can be used). The 1 x 1 mm2 square should contain 16 smaller squares.
- Count all the cells in the four 1 x 1 mm2 corner squares, as well as in the middle square of the 3 x 3 grid. If there are too many or too few cells to count, repeat the procedure, either concentrating or diluting the original cell suspension as appropriate (15-50 cells should overlay a singling 1 mm2 area).
- Calculate the concentration of the cells (cells ml-1) in the standard cell suspension Od0 from the cell count averaged across the five counted squares as follows: Cell count ml-1 = Average cell count square-1 Dilution Factor 104.
- Evaluation of DNA Extraction Efficiency
- Prepare nine sterile 0.5 ml tubes according to Table 2A.
- Dilute the standard cell solution (Od0) to contain about 107 cells ml-1 in a total volume of 20 µl (Od1).
- Add 18 µl of sterile nuclease-free PCR-grade water (NFW) to tubes Od2-Od8 and 20 µl to the blank tube, Od9.
- Transfer 2 µl of the cell solution from Od1 into Od2. Pipette gently to mix and transfer 2 µl from tube Od2 to Od3. Continue diluting the cells in the same manner along the tubes up to and including tube Od8 to obtain a serial dilution of 107-100 cells ml-1.
- In a biological cabinet with circular flow, cut 18 circular pieces of clean, pre-baked filter, using a disinfected 1.12 cm diameter cork borer. Place the filters in pairs into sterile Petri dishes.
- Spike 10 µl from dilutions Od1 to Od8 and from the blank tube Od9 onto each of the paired filters, such that there is one filter pair corresponding to each standard cell concentration.
- Leave the Petri dishes open under the hood and let them dry for 30 min.
- Place each filter piece in a sterile screw-top 2 ml tube and extract the DNA according to the procedure detailed in section 3.4.
- DNA Extraction from the Experimental and Spiked Filters
Note: For DNA extraction from filters, use commercial kits designed for DNA extraction from soil according to the manufacturer's protocol with the following modifications.- For each filter sample (Od1-Od8), prepare a mixture of acid-washed glass beads in a 0.5 ml sterile tube. Use 0.1 g beads having a diameter of 425-600 µm and 0.3 g beads having a diameter of ≤106 µm.
- In a biological cabinet with circular flow, cut three circular pieces from randomly-selected locations on each filter sample using a disinfected 1.12 cm diameter cork borer, and place them in screw-top 2 ml tubes.
- Add the glass bead mixture to the tubes.
- Add the cell-lysis buffer supplied with the extraction kit to each tube in the biological cabinet, with the amount indicated in the manufacturer’s protocol.
- Prepare a bucket of ice and disrupt the cells mechanically using a bead beater.
- Beat the beads for 1 min and then place them on ice for 1 min. Repeat five times.
- Proceed with the kit supplier's protocol from the post-lysis step.
- After elution with the supplied elution buffer (contains 10 mM Tris buffer) or NFW, reload the eluted sample through the column again to improve DNA yield.
Note: If not analyzed on the same day as the DNA extraction, samples can be stored at -20 °C until analysis.
| A- Preparation of Standard Cell Dilution Series | ||||
| Tube | Final cell concentration (cell ml-1) | Standard cell volume (ml) | Nuclease free water volume (ml) | Total volume (ml) |
| Od0 | determine with Hemocytometer counting chamber | |||
| Od1 | should be eluted to the range of 107 cells ml-1 | 20 | ||
| Od2 | 10-1 Od1 | 2 of Od1 | 18 | 20 |
| Od3 | 10-2 Od1 | 2 of Od2 | 18 | 20 |
| Od4 | 10-3 Od1 | 2 of Od3 | 18 | 20 |
| Od5 | 10-4 Od1 | 2 of Od4 | 18 | 20 |
| Od6 | 10-5 Od1 | 2 of Od5 | 18 | 20 |
| Od7 | 10-6 Od1 | 2 of Od6 | 18 | 20 |
| Od8 | 10-7 Od1 | 2 of Od7 | 18 | 20 |
| Blank | 0 | 0 | 20 | 20 |
| B- Preparation of DNA Standard Curve | ||||
| Tube | Final DNA concentration (mg ml-1) | Standard DNA volume (ml) | Nuclease free water volume (ml) | Total volume (ml) |
| Dd0 | determine with NanoDrop | |||
| Dd1 | should be eluted to the range of 101 mg ml-1 | 20 | ||
| Dd2 | 10-1 Dd1 | 2 of Dd1 | 18 | 20 |
| Dd3 | 10-2 Dd1 | 2 of Dd2 | 18 | 20 |
| Dd4 | 10-3 Dd1 | 2 of Dd3 | 18 | 20 |
| Dd5 | 10-4 Dd1 | 2 of Dd4 | 18 | 20 |
| Dd6 | 10-5 Dd1 | 2 of Dd5 | 18 | 20 |
| Dd7 | 10-6 Dd1 | 2 of Dd6 | 18 | 20 |
| Dd8 | 10-7 Dd1 | 2 of Dd7 | 18 | 20 |
| Blank | 0 | 0 | 20 | 20 |
Table 2: DNA Standard Curve. Standard microorganism cell dilution series (A) detailed for the standard cell volume, NFW volume, and the total volume in each dilution tube. DNA standard curve preparation (B), detailed for the standard DNA volume, NFW volume, and the total volume in each dilution tube.
- DNA Standard Curve Preparation
- Extract DNA directly from a standard sample of the microorganism of interest (Dd0) using the same procedure as described for the experimental filters in step 3.4.
- Use a spectrophotometer to determine the DNA concentration of the standard DNA sample (Dd0) at 260 nm.
- Prepare nine sterile 0.5 ml tubes from which to prepare a DNA standard curve, as shown in Table 2B.
- If necessary, dilute the standard DNA solution Dd0 to within a concentration range of 1-10 µg ml-1 in the first tube (Dd1) in a total volume of 20 µl.
- Add 18 µl of NFW to tubes Dd2-Dd8 and 20 µl to the blank tube.
- Transfer 2 µl of the standard DNA solution from Dd1 into Dd2. Pipet gently to mix and then transfer 2 µl from Dd2 to Dd3. Continue diluting the DNA in the same manner along the tubes to achieve a serial dilution of 100-10-7 in tubes Dd2-Dd8 together with a blank tube containing only NFW.
Note: If not analyzed on the same day of extraction, store samples at -20 °C until analysis.
- Quantitative-PCR Analysis
- Switch on the q-PCR instrument in advance to warm up.
- In a new program file, insert the details for the q-PCR run in the instrument-operating software as detailed in Table 3.
Note: Different instruments have different optimal timing for each thermal cycle step. This input is specified in the instrument's manual. - Disinfect the work surface with surface DNA-decontaminant and work only with sterile tubes, tips, and reagents.
- Prepare a bucket of ice next to the working bench. Store Taq-polymerase mix on ice.
- Calculate the total number of reaction wells that will be occupied in the plate. Make theoretical allowance for extra reactions (5%) and multiply the enlarged number of reactions by the volume per reaction to calculate the total reaction volume.
- Prepare the reaction mixed pool, starting from NFW, primers, probe, and lastly the Taq polymerase mix. See example for the volume calculations for the mixed pool in Table 4.
- Store the reaction mix in the dark and on ice until use.
- Place 1 µl of standard DNA, sample DNA, or NFW (as a negative control) inside the wells in the q-PCR microplate in triplicates.
- Ensure that the plate is in a plate holder to prevent it from touching the working surface and to keep it clean.
- Add 9 µl of the mixed pool (Table 4) into each reaction well.
- Cover the plate with optical adhesive film and seal it tightly from all sides.
- Spin down the plate in a centrifuge with plate buckets for 1 min at 1,000 x g before placing it in the q-PCR device.
- Place the plate into the instrument and activate the running program.
| Parameter | Details | Comments |
| Detection method | Quantitative hydrolysis probe | |
| Thermal cycling conditions | ||
| Initial denaturation and enzyme activation | 95 °C for 10 min | |
| Denaturation | 95 °C for 15 sec | repeat 45 times |
| Annealing and extension | 60 °C for 60 sec | |
| Plate array | ||
| Standard curve | Dd1 – Dd8 | 3 repeats per dilution in 1-8 wells at the top 3 rows |
| Non-template control (NTC) | Nuclease free water (NFW) | 3 repeats in the 9th well at the top 3 rows. |
| Analyzed samples | DNA extracted from filters | 3 repeats per sample at the remaining wells. |
| Primer set | per each well | |
| sample volume | 10 ml |
Table 3: Details for q-PCR operating software. Details of the analysis parameters to be entered into the q-PCR program file.
| Reagent | Volume per reaction (ml) | Number of reactions | Allowance for error (5%) | Total volume mix (ml) |
| Taq polymerase mix | 5 | 50 | 52.5 | 262.5 |
| F primer (10 mM) | 0.5 | 50 | 52.5 | 26.25 |
| R primer (10 mM) | 0.5 | 50 | 52.5 | 26.25 |
| Nuclease free water | 3 | 50 | 52.5 | 157.5 |
| DNA – 1 ml will be added directly into the target wells in the 96 plate. | ||||
Table 4: Quantitative-PCR Reaction Mix Calculation. Volume per reaction, number of reactions, allowance for error, and the total calculated volume to be added into the reaction mix per reagent are detailed.
Representative Results
It is common to study atmospheric aerosols using "off-line" analyses of sampled filters (see Figure 2).32 Chemical analyses of the sampled matter include organic (e.g. protein, hydrocarbon molecules, saccharides) and inorganic (e.g. metals, salts) content. Biological analyses include viable and non-viable microorganism content, species identification using a DNA approach or microscopy, as well as DNA-based quantification.
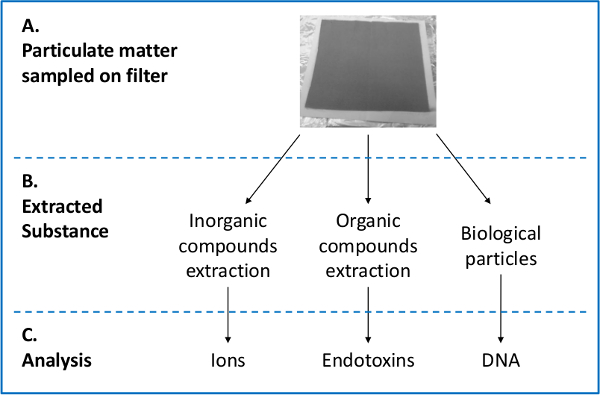
Figure 2: Filer sample analysisflow chart. Filter after air-sampling (A), preparation of the filter for downstream analyses (B), and analysis of the sample components (C).
For the endotoxin extraction, filter sub-samples (1.12 cm2) are shaken in 1 ml PFW for 60 min at room temperature. The samples are then centrifuged for 10 min at 375 x g. Different centrifugation speeds can result in different extraction efficiencies, as indicated in Figure 3A.
A preliminary test for extraction efficiency should be conducted for the specific filter chosen and protocol performed. In Figure 3B, higher extraction efficiencies are obtained from spiking clean compared with sampled quartz filters, and both of these efficiencies are lower than that achieved via direct detection of the standard solution. As discussed elsewhere, the nature of the ambient aerosols sampled on the filter can also affect the endotoxin extraction efficiency.14,33
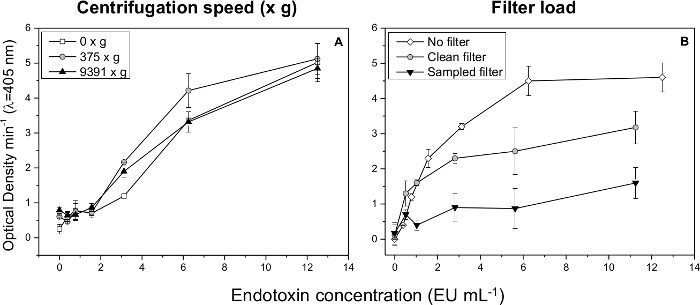
Figure 3: Endotoxin extraction efficiency. The effect of centrifugation speed (A) and filter load (B) on endotoxin extraction efficiency. Error bars represent standard deviation. Please click here to view a larger version of this figure.
The reaction for endotoxin detection involves a 1:1 ratio of LAL reagent to the endotoxin standard, the sample-extracted endotoxins, or the blanks. It is recommended to use triplicates for sample analysis. However, for standard curves, duplicates are sufficient. When the absorption readings (at 405 nm) are completed, the resulting optical density (OD) is analyzed after exporting it into a data-analysis spreadsheet program for further analysis of the data.
Extraction of DNA from the air-sampled filter is performed directly from portions of sub-sampled filters. DNA extraction is performed using a commercial soil-sample DNA isolation kit, with some modifications to increase the DNA yield. Thus, glass beads are added to agitate the filters in a bead-beater to improve the release of DNA into the solution. To prevent sample heating (which may lead to denaturation of the DNA), this step is divided into five 1 min intervals, with the tube cooled on ice between intervals. The extraction can then continue according to the manufacturer's procedure.
As in the endotoxin extraction, it is important to perform a preliminary analysis of the DNA extraction efficiency under the specific experimental conditions. This is done by spiking a known amount of a standard organism cell solution onto a piece cut from the filter, following the DNA extraction protocol described above.34
The concentration of the target microorganism extracted from the sampled aerosols is determined using q-PCR. Here the hydrolysis probe technique is used, which has the advantage of high specificity, because of the additional probe bound to the template DNA.35 The SYBR green method can also be applied for this purpose.
Calibration curves are derived from DNA extracted from the chosen organisms of interest, using the extraction method described above. A mixed pool of the reaction compounds, excluding the DNA sample, standard, or control, is prepared and kept in the dark and on ice until required. Then, an aliquot of the mixed pool is added into each well in a 96 microwell optical plate. The DNA standard, sample, or control is added after the reaction mix according to the plate array. After all reagents have been inserted into the plate, it is sealed with an optical adhesive film and spun down to collect all droplets at the bottom of the wells. The plate is than inserted into the q-PCR instrument for thermal cycle amplification and signal detection.
The output data consist of PCR Ct values, which are defined as the cycle number at which the amplified DNA amount reaches the threshold level. Using the calibration curve values, the concentration of target DNA can be obtained (see Figure 4).36 The microorganism's cell concentrations can be calculated from the Ct values of the extraction efficiency compared with a hemocytometer count of the standard organism solution.37
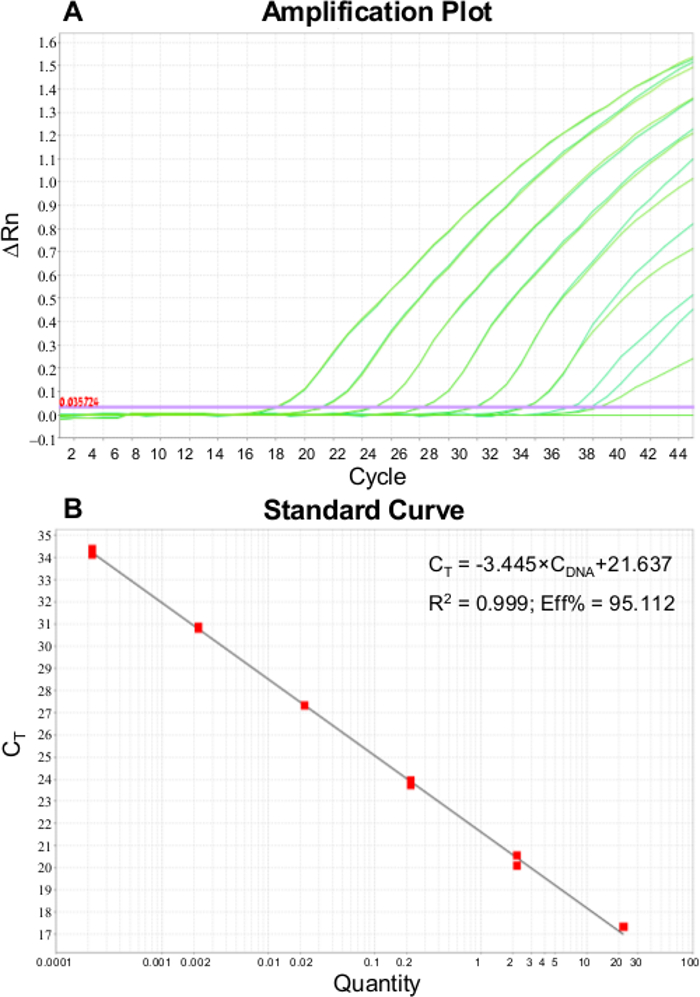
Figure 4: Quantitative-PCR amplification plot. (A) and calibration curve ranging between 2 x 10-4-2 x 102 (B) performed for representative gram negative bacterial DNA. Please click here to view a larger version of this figure.
Discussion
This work describes extraction and detection methods for quantifying both endotoxins and DNA present in aerosol samples collected on filters. The methods require accurate routines and can be performed easily as long as the experimentalist adheres to a few essential and important points discussed here.
For the endotoxin detection step, note that the lysate solution is quite viscous and tends to produce bubbles upon pipetting. It is difficult to remove thee bubbles, and they lead to changes in the microplate-reader values. Therefore, it is important to first place the sample in the plate and only after ensuring that all the bubbles disappeared, continue with the addition of the lysate solution. A helpful technique to prevent bubble formation in this step is to work slowly with the pipette, and not to press it all the way when draining the lysate into the wells. As a reaction start immediately once the lysate is being added to the endotoxin extract, it is essential to start the reading as soon as possible. To shorten the lysate addition step, a multichannel pipette can be used. Note that in some kits and protocols there is an additional pre-incubation step of the samples at 37 °C for 10 min, after it is placed in the micro titter plate.29 Only after this step, the lysate can be added.
An important point for environmental sample extraction of endotoxins is the presence of inhibitors in the samples (e.g. in high pollution air sample). In this case, dilution becomes critical, and can prevent false enhancement, typically observed for non-diluted sample.38
Similarly, there are several points of importance to ensure the success and reliability of the q-PCR detection. 1) As the volume of the solutions is very small, if they are not placed at the bottom of the well, the water content in the DNA drop could be reduced by evaporation while arranging the samples on the plate. To prevent this problem, place the DNA sample at the bottom of the well, cool the plate by placing it on ice or another cooling platform, and prepare everything in advance to quicken this step as much as possible. 2) When preparing the q-PCR reaction mix (Table 4), cover the tube with aluminum foil to prevent photo-degradation. 3) The present experiment describes the preparation of a 10 µl reaction. Nevertheless, in some q-PCR instruments, the reaction volume is 20 µl and then the volume of each reagent is multiplied accordingly. 4) The optimal thermal cycle conditions may change for different primers and polymerase mixes.39 For example, the annealing temperature should be set at around the melting temperature of the primers, and is determined empirically as the temperature at which amplification occurs most efficiently. For this reason it is also important to prepare a calibration curve for each analyzed q-PCR plate, to ensure that the quantification is accurate. Calibration curves as well as standard-spiked filters serve also as a positive control, to ensure that no inhibition occurs. To avoid inhibition, it is recommended to use extraction kits designed to remove inhibitors from environmental samples. 5) The number of thermal cycles in this experiment was set to the maximum number, 45, in order to increase sensitivity as the DNA concentrations from the filter extracts were very low. 6) Ensure that the Cycles to Threshold (Cτ) values for all DNA samples are within the dynamic range of the calibration curve. Also ensure that any amplification of a negative control sample is at least eight cycles greater than that of the DNA samples. 7) If working with the SYBR Green reagent, make sure that there are no multiple melting temperature peaks per reaction.
For turbidimetric test, two endotoxin quantification methods are available: the kinetic method and the endpoint chromogenic method. In the kinetic approach, as performed in this protocol, the time required for the sample to reach maximal absorbance is measured. Here, a shorter reaction interval indicates a higher endotoxin concentration in the sample. In the endpoint chromogenic method, light absorption is measured after a defined incubation time to determine the endotoxin concentration. Both methods require a standard calibration curve for quantification.40
Although the precision and accuracy of the endotoxin extraction described above is very high,14 the extraction efficiency could be improved. It is possible that a different type of filter or the addition of a detergent (such as Polysorbate 20) to the extraction procedure could improve the yield of endotoxin extracted from the filter. Endotoxin extraction efficiency may also be influenced by different particles sampled in parallel on the filter.33 For our experimental setup, the method's limit of detection is estimated to be 0.001 EU m−3.14
Techniques for quantifying the DNA extract that could serve as an alternative to q-PCR include cloning approach and digital droplet PCR (DD-PCR). The advantages of q-PCR over the cloning approach for species identification are its greater sensitivity and that it requires a lower DNA concentration for the initial amplification step. In addition, unlike q-PCR, the cloning method, which is labor-intensive and time-consuming, is not quantitative. An alternative method for the quantification of the DNA extract is DD-PCR.19 The advantage of this technique is that it does not require a calibration curve, as the detection is based on a single DNA strain in each droplet. However, for environmental samples, no reliable method exists so far for droplet-detection of DNA extracted from filters.
The efficiency with which DNA is extracted from the filter, as well as the accuracy and precision of the DNA measurements, can be influenced by factors such as filter type, target microorganisms, and instrumentation.34,41 Therefore it should be defined prior to the sample analysis. In our experimental setup, the limit of detection can reach down to 3.5 genome m-3.14
To conclude, the method described here is applicable for the identification and quantification of air-sampled biological species from both anthropogenic and natural sources. Precise use of the appropriate assay enables valuable investigations of complex environmental questions to be undertaken.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Dr. Yoav Barak from the Chemistry Faculty, Weizmann Institute, for support and advice. This study was supported by the Israel Science Foundation (grant # 913/12), and by the Minerva Foundation with funding from the Federal German Ministry for Education and Research.
Materials
| Filter sampling | |||
| HiVol 3000 – High Air Volume Sampler | Ecotech | ||
| Quartz Microfiber Filters | Whatman | 1851-865 | 203mm X 254mm |
| ELF – Laboratory Chamber Furnaces | Carbolite | ELF 11series | |
| Aluminum foil | Opal | ||
| Name | Company | Catalog Number | Comments |
| Endotoxin | |||
| Ethanol | Sigma Aldrich | 16368 | Laboratory Reagent, 96% |
| Airstream Class II Biological Safety Cabinet AC-4E1 | ESCO | 10011712 | |
| Pyrotell -T | Associates of Cape Cod, Inc. | T0051 | |
| Control Standard Endotoxin | Associates of Cape Cod, Inc. | E0005-1 | Escherichia coli O113:H10, 0.5 µg/vial 1 Pack |
| LAL Reagent Water | Associates of Cape Cod, Inc. | W0051 | |
| 10 mL sterile syringe with Luer-Lok Tip | Becton-Dickinson & Co. | 309605 | |
| BD Precisionglide syringe needle | Becton-Dickinson & Co. | 305129 | Sterile |
| Parafilm-M sealing tape | Parafilm | P7543 | Sigma catalog number |
| Microtubes | Axigen | MCT-200-C | 2ml, pyrogen free |
| 1.12 cm diameter Cork Borer | Boekel Scientific | 1601 BD Series – Steel | Part of a cork borer set containing borers with various diameters. |
| 50 mm Petri Dish | Miniplast Ein-Shemer | 72050-01 | Aseptic |
| Vortex Genie 2 | Scientific Industries, inc. | SI-0297 | |
| Microcentrifuge 5415 D | Eppendorf | 22621408 | |
| TC MicroWell 96 F SI w/lid | Nunc | 167008 | Flat bottom wells (with lid (individually wrapped)), sterile, pyrogen free |
| Synergy HT Multi-Detection Microplate Reader | Biotek | 7091000 | |
| Name | Company | Catalog Number | Comments |
| DNA | |||
| DNA away | Sigma Aldrich | 7010 | |
| Standard DNA of the microbial species of interest | ATCC or other culture collection | Either the appropriate microbial strain for DNA extraction or the extracted DNA | |
| Neubauer-improved | Marienfeld | 640030 | hemocytometer |
| TE buffer, Low EDTA | Life Technologies | 12090-015 | 10 mM Tris-HCl (pH 8.0) 0.1 mM EDTA |
| Nuclease-free PCR-grade water | Sigma Aldrich | 3315959001 | |
| PCR primers | Sigma Aldrich | Targets the microbial species of interest | |
| Dual-Labeled Probes | Sigma Aldrich | Targets the microbial species of interest | |
| Screw cap tubes | Axigen | ST-200-SS | 2 mL |
| PowerSoil DNA extraction kit | Mo Bio Laboratories | 12888-100 | |
| Glass beads, acid-washed 425-600 Microns | Sigma Aldrich | G8772-100G | |
| Glass beads, acid-washed <106 microns | Sigma Aldrich | G4649-100G | |
| PowerSoil Solution C1 | Mo Bio Laboratories | 12888-100-1 | Cell lysis buffer , Power soil Kit |
| Magic Touch ice bucket | Bel-Art | 18848-4001 | |
| Mini-Beadbeater-16 | BioSpec | 607EUR | |
| StepOnePlus Real-Time PCR System | Applied Biosystems | 4376600 | |
| Fast SYBR Green Master Mix | Applied Biosystems | 4385612 | |
| TaqMan Gene Expression Master Mix | Applied Biosystems | 4370048 | |
| MicroAmp Fast Optical 96-Well Reaction Plate with Barcode, 0.1 mL | Applied Biosystems | 4346906 | |
| MicroAmp Splash-Free 96-Well Base | Applied Biosystems | 4312063 | |
| MicroAmp Optical Adhesive Film | Applied Biosystems | 4311971 | |
| Centrifuge 5810 R | Eppendorf | 5811 000.010 | Rotor A-4-62 with MTP buckets |
References
- Lodge, J. P. J. . Methods of Air Sampling and Analysis. , (1988).
- Costa, V., et al. Characteristics of carbonaceous aerosols in Emilia-Romagna (Northern Italy) based on two fall/winter field campaigns. Atmos. Res. , (2015).
- Brent, L. C. . Development, enhancement, and evaluation of aircraft measurement techniques for national ambient air quality standard criteria pollutants [dissertation]. , (2014).
- Okuda, T., Schauer, J. J., Shafer, M. M. Improved methods for elemental analysis of atmospheric aerosols for evaluating human health impacts of aerosols in East Asia. Atmos. Environ. 97, 552-555 (2014).
- Hospodsky, D., et al. Characterizing airborne fungal and bacterial concentrations and emission rates in six occupied children’s classrooms. Indoor air. , (2014).
- Bottos, E., Woo, A., Zawar-Reza, P., Pointing, S., Cary, S. Airborne Bacterial Populations Above Desert Soils of the McMurdo Dry Valleys, Antarctica. Microb. Ecol. 67, 120-128 (2014).
- Ovadnevaite, J., et al. Submicron NE Atlantic marine aerosol chemical composition and abundance: Seasonal trends and air mass categorization. J. Geophys. Res. Atmos. 119, (2014).
- Lodge, J. ES&T Books: Methods of Air Sampling and Analysis, 3rd ed. Environ. Sci. Technol. 23, 938 (1989).
- Pramod, K., Baron, P. A., Willeke, K. . Aerosol Measurement: Principles, Techniques, and Applications. , (2011).
- Vincent, J. H. . Aerosol Sampling: Science, Standards, Instrumentation and Applications. , (2007).
- Duquenne, P., Marchand, G., Duchaine, C. Measurement of Endotoxins in Bioaerosols at Workplace: A Critical Review of Literature and a Standardization Issue. Ann. Occup. Hyg. 57, 137-172 (2013).
- Okuda, T., Schauer, J. J., Shafer, M. M. Improved methods for elemental analysis of atmospheric aerosols for evaluating human health impacts of aerosols in East Asia. Atmos. Environ. 97, 552-555 (2014).
- Lewtas, J. Air pollution combustion emissions: Characterization of causative agents and mechanisms associated with cancer, reproductive, and cardiovascular effects. Mutat. Res.-Rev. Mutat. 636, 95-133 (2007).
- Lang-Yona, N., Lehahn, Y., Herut, B., Burshtein, N., Rudich, Y. Marine aerosol as a possible source for endotoxins in coastal areas. Sci. Total. Environ. 499, 311-318 (2014).
- Levin, J., Bang, F. B. Clottable protein in Limulus: its localization and kinetics of its coagulation by endotoxin. Thromb. Diath. Haemorrh. 19, 186-197 (1968).
- Heid, C. A., Stevens, J., Livak, K. J., Williams, P. M. Real time quantitative PCR. Genome Res. 6, 986-994 (1996).
- O’Dowd, C. D., de Leeuw, G. Marine aerosol production: a review of the current knowledge. Phil. Trans. R. Soc. A. 365, 1753-1774 (2007).
- O’Dowd, C. D., et al. Biogenically driven organic contribution to marine aerosol. Nature. 431, 676-680 (2004).
- Yang, R., Paparini, A., Monis, P., Ryan, U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol. 44, 1105-1113 (2014).
- Stetzenbach, L. D., Buttner, M. P., Cruz, P. Detection and enumeration of airborne biocontaminants. Curr. Opin. Biotechnol. 15, 170-174 (2004).
- Dannemiller, K. C., Gent, J. F., Leaderer, B. P., Peccia, J. Influence of housing characteristics on bacterial and fungal communities in homes of asthmatic children. Indoor air. , (2015).
- Yamamoto, N., Hospodsky, D., Dannemiller, K. C., Nazaroff, W. W., Peccia, J. Indoor emissions as a primary source of airborne allergenic fungal particles in classrooms. Environ. Sci. Technol. 49, 5098-5106 (2015).
- Yamamoto, N., et al. Particle-size distributions and seasonal diversity of allergenic and pathogenic fungi in outdoor air. ISME J. 6, 1801-1811 (2012).
- Karottki, D. G., et al. Cardiovascular and lung function in relation to outdoor and indoor exposure to fine and ultrafine particulate matter in middle-aged subjects. Environ Int. 73, 372-381 (2014).
- Guy, D., Hodges, N., Hanlon, G. Endotoxins and Depyrogenation. Industrial Pharmaceutical Microbiology: Standards and Controls. , 12.1-12.15 (2003).
- Thorne, P. S., Bartlett, K. H., Phipps, J., Kulhankova, K. Evaluation of Five Extraction Protocols for Quantification of Endotoxin in Metalworking Fluid Aerosol. Ann. Occup. Hyg. 47, 31-36 (2003).
- Thornton, B., Basu, C. Real-time PCR (qPCR) primer design using free online software. Biochem. Mol. Biol. Educ. 39, 145-154 (2011).
- Madigan, M. T., Clark, D. P., Stahl, D., Martinko, J. M. . Brock Biology of Microorganisms. , (2011).
- Watson, J. G., Chow, J. C. . Aerosol Measurement: Principles and Techniques. , 591-613 (2011).
- Mueller-Anneling, L., Avol, E., Peters, J. M., Thorne, P. S. Ambient endotoxin concentrations in PM10 from Southern California. Environ. Health Perspect. 112, 583-588 (2004).
- Hospodsky, D., Yamamoto, N., Peccia, J. Accuracy, Precision, and Method Detection Limits of Quantitative PCR for Airborne Bacteria and Fungi. Appl. Environ. Microbiol. 76, 7004-7012 (2010).
- Kutyavin, I. V., et al. 3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures. Nucleic Acids Res. 28, 655-661 (2000).
- Brankatschk, R., Bodenhausen, N., Zeyer, J., Bürgmann, H. Simple absolute quantification method correcting for quantitative PCR efficiency variations for microbial community samples. Appl. Environ. Microbiol. 78, 4481-4489 (2012).
- Strober, W. Appendix 3B, Trypan Blue Exclusion Test of Cell Viability. Current Protocols in Immunology. , (2001).
- Hollander, A., Heederik, D., Versloot, P., Douwes, J. Inhibition and enhancement in the analysis of airborne endotoxin levels in various occupational environments. Am Ind Hyg Assoc J. 54, 647-653 (1993).
- Kennedy, S. . PCR troubleshooting and optimization : the essential guide. , (2011).
- Joiner, T. J., Kraus, P. F., Kupiec, T. C. Comparison of Endotoxin Testing Methods for Pharmaceutical Products. Int J Pharm Compd. 6, 408-409 (2002).
- Ebentier, D. L., et al. Evaluation of the repeatability and reproducibility of a suite of qPCR-based microbial source tracking methods. Water Res. 47, 6839-6848 (2013).

