Gene Expression Profiling of Infecting Microbes Using a Digital Bar-coding Platform
Summary
We developed a protocol that is fast, sensitive and reproducible for pathogen gene expression profiling during an infection.
Abstract
For most mammalian pathogens, gene expression profiling studies have been limited by technical difficulties to accurately quantify pathogen gene transcripts from infected tissues. Pathogen RNA constitutes a tiny portion of the total RNA isolated from infected tissue samples. Both microarray and RNAseq technologies have difficulties in generating reliable reads for weakly expressed pathogen genes. Mutant pathogen strains with reduced in vivo proliferation pose an even bigger challenge. Here we describe an in vivo gene expression profiling protocol that is very fast, extremely sensitive and highly reproducible. We developed this protocol during our investigation of the fungal pathogen Candida albicans in a murine model of hematogenously disseminated candidiasis. Using this protocol, we have documented time courses of dynamically regulated C. albicans gene expression during kidney infection, and discovered unexpected features of gene expression responses to antifungal drug treatment in vivo.
Introduction
Gene expression dynamics holds vital information about how a cell responds to environmental changes. In the case of an infecting microbe, gene expression data could provide clinically relevant insights on how a pathogen adapts to the infection environment and causes damage to the host. In the past two decades, gene expression studies both in vitro and in vivo have generated large amount of data and laid a foundation for understanding infection biology1-3. However, current profiling technologies including microarray and RNAseq, are not optimal for profiling of pathogen gene expression during infection. Host RNA constitutes an overwhelming portion (usually > 99%) of the total RNA isolated from infected tissue samples. The host RNA contributes to high background on microarrays, and dominates sequence reads from RNAseq. Pathogen cell isolation from infected tissue, in theory, can help to enrich for pathogen RNA, but it poses additional problems: it requires large quantities of infected tissue; the procedure can be tedious; and most importantly, it is difficult to conserve the native state or integrity of RNA during the lengthy process. In order to generate more comprehensive and accurate data on pathogen gene expression during real infections, we set out to develop a protocol that allows reliable and cost-effective quantification of minority RNA species in a mixed RNA population.
NanoString nCounter is a recently developed platform that makes direct multiplexed measurement of gene expression using digital bar-coded probe pairs4, 5. This platform has a sensitivity level comparable to that of QRT-PCR but does not require enzymatic amplification. The technology is not genome-wide, it allows detection of up to 800 different genes in each assay. Therefore, it is critical to select informative genes as probes for profiling. Here we use gene expression profiling study of a major human pathogen, Candida albicans, during an invasive infection as an example, to demonstrate: 1) Technical details of the experimental procedures (protocol), 2) The sensitivity, reproducibility and dynamic range of this method (representative results), and 3) Important considerations for designing and performing experiments based on this protocol (discussion). This protocol can be easily adapted for various pathogens and has been successfully applied in a number of infection models6-9.
Protocol
All animal procedures were approved by the Institutional Animal Care and Use Committee at the Los Angeles Biomedical Research Institute (protocol 011000) and carried out according to the National Institutes of Health (NIH) guidelines for the ethical treatment of animals. The mice were caged in an AAALAC-accredited facility located on the campus of Harbor-UCLA Research and Education Institute. A full-time veterinarian who specializes in laboratory animal medicine oversaw their care. Caging and husbandry was provided according to the guidelines in the US Public Health Service publication "Guide for the Care and Use of Laboratory Animals". Every attempt was made to treat the mice humanely. The survival and health of the mice was monitored three times daily. Obviously sick, lethargic mice were segregated from the group and euthanized to minimize suffering. The mice were euthanized by pentobarbital overdose (210 mg/kg), as recommended by the Panel on Euthanasia of the American Veterinary Medical Association.
1. Preparation of Tissues, Reagents and Instruments
- Use male Balb/c mice weighting 20-22 g for all studies. Inoculate three mice per experimental group intravenously with 1 x 106 yeast-phase C. albicans cells. Sacrifice animals and harvest kidneys at specific time points post-infection. Place each kidney into a screw cap tube, snap freeze in liquid nitrogen, and store at -80 °C for later RNA extraction6.
Note: The animal experiments were carried out as described in detail in Xu et al.6. - Prepare the following reagents and instruments before removing kidneys from the -80 °C freezer:
- Open a total RNA extraction kit. Add 2-mercaptoethanol (1% v/v) to buffer RLT and mix well (prepare 2 ml for each sample).
- Prepare phenol : chloroform : isoamyl alcohol 25:24:1 (need 600 µl for each sample).
- Label M-type homogenization tubes (M-tube) and chill on ice.
- Label 2 ml screw cap tubes, and add approximately 300 µl Zirconia beads to each tube.
- Check instruments are available: tissue dissociator, beadbeater, tabletop centrifuge with adaptors for 50 ml tubes and 96 well plates, photometer.
2. RNA Extraction from Infected Tissues
- Remove tubes containing kidneys from -80 °C freezer and put on ice. Add 1.2 ml buffer RLT with 2-mercaptoethanol to each kidney. Decant kidney with buffer into an M-type homogenization tubes.
- Homogenize the kidney using a tissue dissociator on pre-loaded setting RNA_02.01.
- Centrifuge at 1,000 x g for 1 min in a tabletop centrifuge at RT.
- Transfer 600 µl homogenate to the screw cap tube containing Zirconia beads. Save the remaining homogenate on ice.
- Add 600 µl phenol : chloroform : isoamyl alcohol 25:24:1 to the tubes from the previous step.
- Close the lids tightly and vortex on beadbeater for 3 min in a 4 °C cold room.
- Centrifuge the tubes at 15,000 x g for 5 min in a 4 °C cold room.
- Carefully transfer the aqueous phase to a new 1.5 ml microfuge tube, mix well with equal volume of 70% ethanol, then load onto the spin column (load twice).
- Wash the spin column once with 700 µl buffer RW, followed by twice with 500 µl buffer RPE. Centrifuge one extra minute in a dry collection tube to remove remaining liquid in the spin column. Perform all centrifugation steps at 8,000 x g.
- Elute RNA with 50 µl H2O. Measure RNA concentration at OD260 using a spectrophotometer.
3. Gene Expression Profiling Using Digital Bar-coding
- Thaw reporter codeset (green cap tube) and capture codeset (gray cap tube) on ice.
- Add 130 µl hybridization buffer to the reporter codeset, invert to mix and spin down.
- Add 20 µl of the mix to each of the 12 reaction tubes.
- Add 10 µg of total tissue RNA (in a volume of 5 µl) to each tube. Mix by pipetting.
- Add 5 µl capture codeset to each tube. Mix by flipping the tube followed by a quick spin down.
- Incubate the reactions at 65 °C in a thermal cycler with heated lid O/N (~18 hr).
- Remove one sample cartridge from -20 °C and let it warm to RT.
- Remove two reagent plates from 4 °C and centrifuge at 670 x g for 2 min in a tabletop centrifuge.
- Set up the prep station following step-by-step instructions on the screen, select the high sensitivity option.
- Remove the reactions from the thermal cycler and immediately load on the prep station. Select to run the high sensitivity program (3 hr).
- When the prep station program is complete, remove the cartridge and seal the lanes with a clear tape. Apply mineral oil to the bottom of the cartridge (for generation one nCounter only, later generations do not require this step), then load the cartridge onto the digital analyzer.
- Set up the digital analyzer following step-by-step instructions on the screen, select the high resolution scan option.
- Select the high resolution (600 fields) option, run the scanning program (~4.5 hr).
- When the scanning program is complete, download the results (or choose to receive the results by email), and import raw data into manufacturer's software. The software will automatically check data quality and raise flags if the quality of the data falls out of the normal range. Perform technical adjustment using the built-in function (optional, follow instructions in the software), then export the data as an excel file.
- Normalize the data using one of the following methods: total counts from all genes in the codeset; one or a few internal control genes; geometric mean of highly expressed genes (see discussion).
- Calculate the mean expression values for each gene (if the experiment was done in replicates or triplicates), then calculate the ratio of expression levels among different experimental groups.
- (optional) Analyze and visualize the profiling results by built-in functions in the nSolver software or a clustering program such as MultiExperiment Viewer10.
Representative Results
One main challenge for pathogen gene expression profiling in vivo is to get enough reads from pathogen RNA. Given the low percentage of pathogen transcripts in total RNA, large amount of total RNA (>10 µg) has to be used for each reaction. The platform has a unique advantage in this perspective: it uses both a capture probe and a report probe to increase the specificity, so that the overwhelming amount of host RNA does not cause a significant level of noise. As shown in Figure 1 (adapted from Ref. 6), background raw counts from an uninfected tissue sample (red dots) were all below 10, while raw counts from two infected tissue samples (blue dots) were all above 10. Expression data generated using this protocol are highly reproducible. As shown in Figure 1 (adapted from Ref. 6), raw counts from two biological replicates (blue dots, 48 hr post-infection kidney samples) had very good correlation, with R-square value equals 0.945. The platform also provides sufficient dynamic range to encompass natural biological expression levels (Figure 1, raw counts ranged between 101 and 106).
Using a probe set specifying 248 environmental response genes, we were able to discern two phases of pathogen gene expression (Figure 2, adapted from Ref. 6): an early gene expression response comprises genes with RNA levels significantly different between the inoculum and 12 hr post-infection samples (P<0.05 and >2-fold change in expression), and a late gene expression response comprises genes with RNA levels that are significantly different between the 12 hr time point at 48 hr post-infection samples (P<0.05 and >2-fold change in expression). These results indicate that C. albicans gene expression is dynamically regulated during invasive infection of a mammalian host.
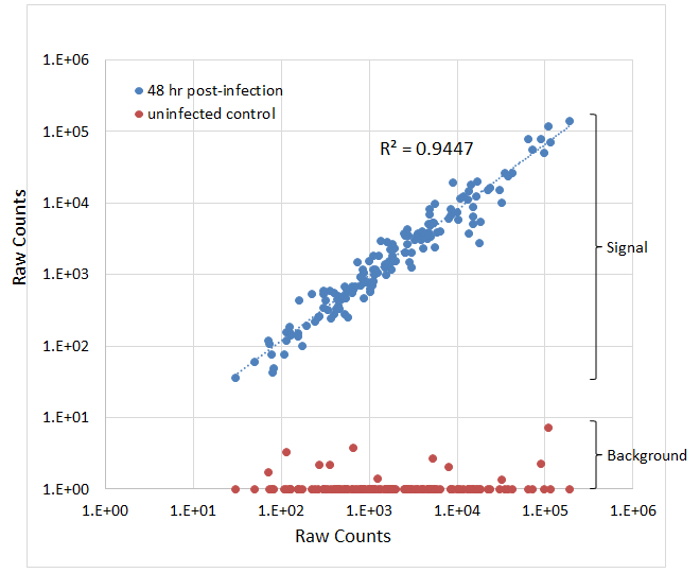
Figure 1. Raw counts from infected and uninfected kidney tissues (adapted from Ref. 6). Probe counts for two infected (48 hr post-infection, blue data points) and one uninfected (red data points) kidney samples are presented as a scatter plot. Please click here to view a larger version of this figure.
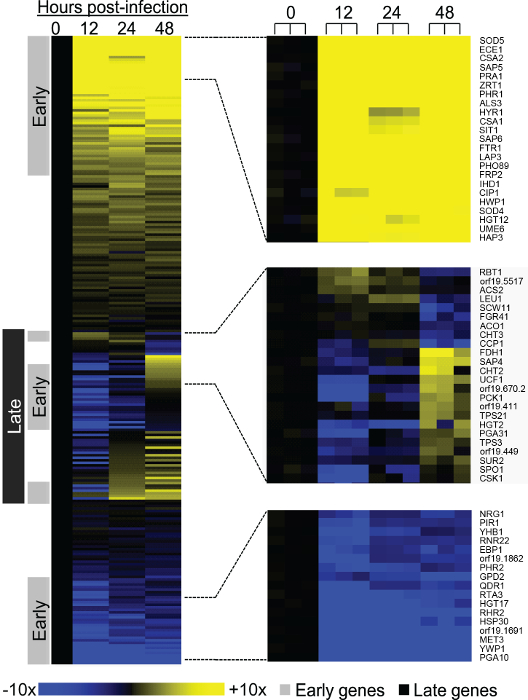
Figure 2. Expression of C. albicans environmentally responsive genes during invasive infection (adapted from Ref. 6). Changes in expression levels during mouse kidney invasion for 248 C. albicans genes are presented in a heat map format. Mean values of biological triplicates are shown for up-regulation (yellow) and down-regulation (blue) of genes at 12, 24, and 48 hr post-infection relative to mean inoculum levels (0 hr). Color saturation represents the extent of the expression change, with full saturation at 10 fold up- or down-regulation. Portions of the heat map are expanded to illustrate representative early up-regulated genes (top), late genes (middle), and early down-regulated genes (bottom). In these portions, individual samples are presented separately to illustrate reproducibility. We define early expression changes as significant differences between the inoculum and 12 hr samples. We define late expression changes as significant differences between the 12 and 48 hr samples. Significance refers to changes of >2 fold and a p-value <0.05. The data for each sample were normalized to RNA levels from control gene TDH3 before mean values were calculated. Our assignment criteria allow some dynamically regulated genes to fall into both the early and late expression classes. Please click here to view a larger version of this figure.
Discussion
This protocol is developed to optimize transcriptional profiling of infecting microbes using a nanoString nCounter platform. The whole procedure, from tissue collection to expression data, requires less than 48 hr. The hands-on time is around 4 hr for 12 samples. One key variable in this protocol is the amount of buffer RLT added to the tissue at the very first step. Too much buffer will dilute the homogenate and lead to lower RNA concentration, while too little buffer may lead to formation of viscous gels after the phenol chloroform extraction step and leave no aqueous phase for RNA recovery. The empirically determined optimal volumes of buffer RLT for mouse tissues (assuming typical sizes) are: kidney (1.2 ml), tongue (1.0 ml) and lung (2.0 ml). For microbial pathogens, especially these grown in filamentous form, the bead-beating step helps to break open thick cell wall to fully release cellular contents. In the elution step, in order to increase the final RNA concentration, the first round eluate can be added back to the column, and elute a second time. Approximately 100 µg of total tissue RNA can be recovered from one RNeasy spin column (~2 µg/µl x 50 µl). If larger amount of RNA is needed, a second prep can be made from the remaining tissue homogenate. Do not dispose of the remaining homogenate until RNA concentration has been measured, just in case.
Depending on the infection model, the inoculum size and the pathogen strain, the percentage of pathogen RNA in total tissue RNA varies from 0%-2%, and typically falls within the 0.05%-0.5% range. For example, 10 µg of total tissue RNA from C. albicans infected kidney could generate raw counts equal to that of 10 ng of pure C. albicans RNA from an in vitro culture (10 ng/10 µg = 0.1%). Because the percentage of pathogen RNA in total tissue RNA varies in a wide range, it is hard to know the quantity of pathogen RNA in a given amount of total RNA. To avoid wasting codeset (for 250 genes x 192 reactions, the cost per reaction is around $200) on samples with pathogen RNA below the detection level, an RNA quality control step may be performed to determine the pathogen RNA level within each sample. This can be done by either Q-RTPCR or a small codeset containing a few housekeeping genes.
Normalization using pathogen genes is a critical step before the expression profiles can be compared among different samples. There are three commonly used methods for normalization. 1. Use total counts from all genes in the codeset. 2. Use one or a few 'housekeeping' genes. 3. Use the geometric mean (Nth root of the product of N numbers) of highly expressed genes. For a large codeset (>100 genes) containing randomly selected probes, using total counts for normalization can be a good choice, because the total counts of a large number of unrelated genes may faithfully reflect the amount of RNA input. For a small codeset (<100 genes) containing probes focused on a specific process (such as hyphal growth, given that hyphal growth genes tend to be co-regulated), choosing one or a few housekeeping genes as control for normalization is essential. TDH3, a robustly expressed metabolic gene, has served well as control for many experiments. The third method is a hybrid of method 1 and 2, with an emphasis for equal contribution from highly expressed genes.
Though the nCounter platform is not genome-wide, it allows quantification of expression level for up to 800 genes in a single assay. Therefore, choosing the most informative genes to analyze is critical for the success of the profiling. Two approaches to select for probes have been used. The first approach is "knowledge based". Information from published expression and functional data is compiled to screen for genes that are of potential interest to the current study, such as the environmental response genes6-9. This approach allows easy comparison of in vivo profiling data to many existing datasets, hence providing context to data interpretation. The second approach is "exploration based". A category of genes in a microbe genome, such as all genes specifying transcription factors, kinases or cell wall proteins are chosen for probes. This approach allows identification of novel virulence factors and discovery of novel regulatory relationships based on the in vivo profiling data6.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported in part by NIH grants R21 DE023311 (APM), R56 AI111836 (APM & SGF), R01 AI054928 (SGF), and R01 DE017088 (SGF).
Materials
| gentleMACS Dissociator | Miltenyl Biotec | 130-093-235 | |
| gentelMACS M tube | Miltenyl Biotec | 130-093-236 | |
| RNeasy Mini Kit | QIAGEN | 74104 | |
| 2-mercaptoethanol | Sigma | M-3148 | |
| Phenol:ChCl3:IAA | Sigma | P-2069 | |
| Zirconia beads | Biospec Products | 11079110zx | |
| Mini-Beadbeater-16 | Biospec Products | 607 | |
| nCounter Analysis system | nanoString Technologies | ||
| nCounter Gene Expression Codesets | nanoString Technologies | ||
| nSolver Analysis tool | nanoString Technologies |
References
- Cairns, T., Minuzzi, F., Bignell, E. The host-infecting fungal transcriptome. FEMS Microbiol Lett. 307 (1), 1-11 (2010).
- Westermann, A. J., Gorski, S. A., Vogel, J. Dual RNA-seq of pathogen and host. Nat Rev Microbiol. 10 (9), 618-630 (2012).
- Creecy, J. P., Conway, T. Quantitative bacterial transcriptomics with RNA-seq. Curr Opin Microbiol. 23C, 133-140 (2015).
- Geiss, G. K., et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol. 26 (3), 317-325 (2008).
- Malkov, V. A., et al. Multiplexed measurements of gene signatures in different analytes using the Nanostring nCounter Assay System. BMC Res Notes. 2, 80 (2009).
- Xu, W., et al. Activation and Alliance of Regulatory Pathways in C. albicans during Mammalian Infection. PLoS Biology. 13, e1002076 (2015).
- O’Meara, T. R., et al. The Cryptococcus neoformans Rim101 Transcription Factor Directly Regulates Genes Required for Adaptation to the Host. Mol Cell Biol. 34 (4), 673-684 (2014).
- Cheng, S., et al. Profiling of Candida albicans Gene Expression During Intra-Abdominal Candidiasis Identifies Biologic Processes Involved in Pathogenesis. J Infect Dis. 208 (9), 1529-1537 (2013).
- Fanning, S., et al. Divergent Targets of Candida albicans Biofilm Regulator Bcr1 in Vitro and In Vivo. Eukaryot Cell. 11 (7), 896-904 (2012).
- Saeed, A. I., et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 34 (2), 374-378 (2003).

