Isolation of Perivascular Multipotent Precursor Cell Populations from Human Cardiac Tissue
Summary
Human cardiac tissue harbours multipotent perivascular precursor cell populations that may be suitable for myocardial regeneration. The technique described here allows for the simultaneous isolation and purification of two multipotent stromal cell populations associated with native blood vessels, i.e. CD146+CD34– pericytes and CD34+CD146– adventitial cells, from the human myocardium.
Abstract
Multipotent mesenchymal stem/stromal cells (MSC) were conventionally isolated, through their plastic adherence, from primary tissue digests whilst their anatomical tissue location remained unclear. The recent discovery of defined perivascular and MSC cell marker expression by perivascular cells in multiple tissues by our group and other researchers has provided an opportunity to prospectively isolate and purify specific homogenous subpopulations of multipotent perivascular precursor cells. We have previously demonstrated the use of fluorescent activated cell sorting (FACS) to purify microvascular CD146+CD34– pericytes and vascular CD34+CD146– adventitial cells from human skeletal muscle. Herein we describe a method to simultaneously isolate these two perivascular cell subsets from human myocardium by FACS, based on the expression of a defined set of cell surface markers for positive and negative selections. This method thus makes available two specific subpopulations of multipotent cardiac MSC-like precursor cells for use in basic research and/or therapeutic investigations.
Introduction
The heart has long been considered a post-mitotic organ. However, recent studies have demonstrated the presence of limited cardiomyocyte turnover in adult human hearts1. Native stem/progenitor cells with cardiomyocyte differentiation potential have also been identified within the myocardium in adult rodent and human hearts, including Sca-1+, c-kit+, cardiosphere-forming, and most recently, perivascular precursor cells2,3. These cells represent attractive candidates for therapies aimed at enhancing cardiac repair/regeneration through cell transplantation or stimulation of in-situ proliferation.
Mesenchymal stem/stromal cells (MSC) have been isolated from almost every human tissue4,5 Clinical trials of the therapeutic applications of MSC have been carried out for multiple pathological conditions such as cardiovascular repair6, graft-versus-host-disease7, and liver cirrhosis8. Beneficial effects have been attributed to the ability of MSCs to: home to sites of inflammation9; differentiate into different cell types10; secrete pro-reparative molecules11; and modulate host immune responses12. The isolation of MSCs has traditionally relied on their preferential adherence to plastic substrates. However, the resulting population of cells is typically markedly heterogenous13. By using fluorescent activated cell sorting (FACS) with a combination of key perivascular cell markers, we have been able to isolate and purify a multipotent MSC-like precursor population (CD146+/CD31–/CD34–/CD45–/CD56–) from multiple human tissues including adult skeletal muscle and white fat14.
Perivascular cell populations in various non-cardiac tissues have been shown to have stem/progenitor cell properties and are being investigated for clinical use in the cardiovascular setting. Pericytes, one of the most well-known perivascular cell subsets, are a heterogeneous population that play several pathophysiological roles including in the development of new vessels15, the regulation of blood pressure16, and maintenance of vascular integrity17,18. As shown in multiple tissues, specific subsets of pericytes natively express MSC antigens and sustain their MSC-like phenotypes in primary culture after FACS purification14. Moreover, these cells stably maintain their long-term phenotypes within culture and exhibit multi-lineage differentiation potential, similar to MSCs19,20. These results suggest that pericytes are one of the origins of the elusive MSC14. The therapeutic potential of pericytes has been demonstrated with a reduction in myocardial scarring and enhanced cardiac function following transplantation into ischemically injured hearts21. Recently, we successfully purified pericytes from the human myocardium and demonstrated their MSC-like phenotypes and multipotency (adipogenesis, chondrogenesis and osteogenesis) with the absence of skeletal myogenesis3. In addition, myocardial pericytes exhibited differential cardiomyogenic potential and angiogenic capacities when compared with counterparts purified from other organs.
A second population of multipotent perivascular stem/progenitor cells, the adventitial cell, has been isolated from human saphenous veins on the basis of positive CD34 expression22. Venous adventitial cells have been shown to have clonogenic potential, mesodermal differentiation capacity and proangiogenic potential in vitro. Transplantation of these cells into the ischemically injured hearts of mice resulted in a reduction in interstitial fibrosis, an increase in angiogenesis and myocardial blood flow, reduced ventricular dilation, and increased cardiac ejection fraction23. Interestingly, adipose adventitial cells have been shown to lose CD34 expression and upregulate CD146 expression in culture in response to angiopoietin II treatment, suggesting the adoption of a pericyte phenotype with stimulation24. Within the heart, however, the adventitial cell population has not yet been prospectively purified by FACS and/or well characterized. Utilizing the cell isolation procedures described in the following sections, we are currently characterizing myocardial adventitial cells and investigating their potential for regenerative applications.
Herein we describe a method to isolate and purify two subpopulations of perivascular stem/progenitor cells from human fetal or adult myocardium. This prospective cell isolation method will enable researchers to obtain isogenic perivascular stem/progenitor cell subsets from human heart biopsies for comparative studies and further explore their therapeutic potential in various cardiac pathological conditions.
Protocol
1. Processing of Human Cardiac Sample
- Ensure that all fluids, containers, instruments, and the dedicated operational area are sterile.
- Place the cardiac tissue sample (procured by the tissue bank or surgical team) in storage medium composed of chilled Dulbecco's modified Eagle medium (DMEM) containing 20% fetal bovine serum (FBS) and 1% penicillin-streptomycin (P/S) on ice for transportation3.
- Remove the cardiac sample from the storage medium and wash with a washing medium composed of phosphate-buffered saline (PBS) supplemented with 2% FBS and 1% P/S. When handling the sample, use fine tipped forceps to grasp large vessels or pericardium and minimise crushing of the myocardium.
- Submerge the sample in washing medium in a petri dish and remove the pericardium, endocardium and large blood vessels with sterilized iris scissors and fine-tipped forceps as described3.
- Cut the remaining myocardium into small pieces of approximately 1 mm3 using a single side razor blade or a pair of sharp iris scissors. Note: Highest cell yields will be obtained if samples are processed immediately. If a delay is unavoidable, storage of the processed cardiac tissue in the fresh storage medium (>5 ml per 1 cm3 tissue) at 4 °C for up to 72 hr is possible, however presumably with an associated decline in the cell yield.
2. Digestion of the Tissue and Isolation of Cells
- Freshly make up the digestion solution comprising 1.5 ml each of collagenase I, collagenase II, and collagenase IV (all at 0.5 mg/ml in DMEM) and warm to 37 °C in a waterbath.
Note:Volume and concentration of collagenases are suitable for samples of approximately 1 cm3. - Filter the suspended tissue pieces through a 100 µm strainer and wash twice with PBS. Transfer the tissue pieces to a sterile 30 ml container with a tightly sealed lid.
Note: Short wide pots rather than tall narrow tubes give better results. Alternatively, put tissue pieces in a sterile 50 ml tube and place horizontally if a 30 ml container is not available. - Add all the digestion solutions (4.5 ml total) and place the container within a sealed plastic bag in a 37 °C shaking water bath set to 120 rpm. Alternatively, seal the container with paraffin film and place in a heated orbital shaker set to 120 rpm.
- After 15 min, remove and invert the pot three times before replacing for a further 15 min. Remove and quench the collagenases with 5 ml of DMEM supplemented with 20% FBS and 1% P/S.
- Triturate the digest by gently pipetting ten to twenty times with a 10 ml serological pipette to break up any tissue clumps. Filter the suspension sequentially through 100 µm, 70 µm and finally 40 µm cell strainers to remove undigested materials and obtain a single cell suspension.
Note: Do not rinse between cell strainers to avoid cell debrisgenerated by the enzyme digestion process. - Centrifuge the cell suspension at 200 x g for 4 min, carefully decant the supernatant and re-suspend the pellet in 1 ml of red cell lysing buffer for 2 min at room temperature before diluting with 4 ml of washing medium.
- Repeat the centrifugation step and re-suspend the pellet in 1 ml of washing medium. Mix well and take 10 µl of the cell suspension for cell counting.
- Mix 10 µl of the cell suspension with 10 µl of Trypan blue stain and place 10 µl of the mixture on a standard hemocytometer for cell counting. Count the number of bright, round cells present in the four corner squares (each subdivided into sixteen small squares) and divide by 4 to get the mean per corner square. Multiply the mean by 2 to account for the stain dilution factor and then by 104 to obtain the total number of cells per 1 ml of sample.
3. Cell Labelling and Sorting
- Add 50 µl of mouse serum to the cell suspension and incubate at 4 °C for 20 min to block non-specific antibody binding.
- Centrifuge the cell suspension with mouse serum at 200 x g for 4 min, remove supernatant, and re-suspend in washing medium at a concentration of 1 x 106 cells per 100 µl.
- Aliquot 50 µl each of the cell suspension for the isotype and unstained controls into two polystyrene round bottom flow cytometry tubes then place the remaining volume into a third tube for multi-color staining.
- Add CD34-PE, CD45-APC-Cy7, CD56-PE-Cy7, CD144-PerCP-Cy5.5, and CD146-AF647 antibodies (all 1:100) to the single cell suspension for multi-color staining. For the isotype control, add equivalent volumes of PE-, APC-Cy7-, PE-Cy7-, PerCP-Cy5.5- and AF647-conjugated isotype antibodies. Gently pipette to mix the antibodies with the cells and incubate all tubes at 4 °C for 20 min in the dark.
- Prepare compensation control beads by adding one drop of positive beads to 100 µl of washing medium in five polystyrene round bottom flow-cytometry tubes. Add CD34-PE, CD45-APC-Cy7, CD56-PE-Cy7, CD144-PerCP-Cy5.5, and CD146-AF647 antibodies (all 1:100) individually to each of the 5 tubes. Incubate all tubes at 4 °C for 20 min in the dark.
- During the incubation, prepare cell collection tubes each with 500 µl of Endothelial Growth Medium-2 (EGM-2) culture medium and place at 4 °C.
- Following antibody incubation, add 5 ml of washing medium to wash cells and beads in order to remove unbound antibody. Centrifuge all tubes at 200 x g for 4 min. Repeat the washing and centrifugation step. Carefully decant the supernatant and pipette gently when re-suspending cells.
- Re-suspend in washing medium at a concentration of approximately 1 x 106 cells per 250 µl. Re-suspend the beads in 100 µl of washing medium.
- Transport all cell and bead suspensions to the cell sorter on ice in the dark. Add 1 drop of negative beads to the positive bead compensation control tubes and use these to set the fluorescence compensation.
- Run unstained control cells according to manufacturer's instructions to establish the background fluorescence and set the voltages to the following: FSC 150V; SSC 290V; V450/50 350V; V710/50 620V; B530/30 400V; B710/50 530V; YG780/60 460; R670/30 480V; R780/60 460V. Run isotypes controls according to manufacturer's instructions to establish the background fluorescence thresholds related to non-specific binding. Run the multi-color stained sample and collect the pericyte and adventitial cell populations into the collection tubes (Figure 1).
NOTE: Optimal viable cell yields are approximately 3% of the total live cell dissociation for pericytes and 4% for adventitial cells.
4. Cell Culture
- Select suitable size culture plates according to the outcome of the FACS sorting. Add 100 µl of sterile 0.2% gelatin solution per cm2 of growth area and agitate manually to coat the entire wells. Incubate plates at 4 °C for 10 min and remove gelatin solution completely. Keep collected cells on ice whilst preparing culture plates.
Note: Freshly sorted pericytes and adventitial cells grow well when seeded at a density of 30,000 to 40,000 cells/cm2 on gelatin-coated culture surface. - Centrifuge freshly collected cells at 200 x g for 4 min and gently re-suspend the cell pellet in an appropriate amount of EGM-2.Seed the cells on gelatin-coated plates.
Note: The actual number of cells to be seeded per well and the amount of EGM-2 to be added depend on the plate selection. For example, 0.5 ml and 1 ml EGM-2 are needed per well for 48- and 24-well plates respectively. - Exchange EGM-2 for perivascular cell growth medium (DMEM supplemented with 20% FBS and 1% P/S) for both pericytes and adventitial cells once cells have settled and adhered to the plate (after at least 72 hr). Change the media every 72 hr from then on. Carry out initial and all subsequent incubations in 5% CO2 and at 37 °C.
- Dissociate cells using 0.05% Trypsin EDTA when pericytes and adventitial cells reach 80 to 90% confluence. Quench with 20% FBS in PBS, centrifuge at 200 x g for 4 min, re-suspend in perivascular cell growth medium, and then passage the cells at a 1:3 ratio onto uncoated polystyrene culture plates. From Passage 2 onwards, passage cells at a 1:5 ratio (or at approximately 7,000 cells/cm2) to uncoated polystyrene culture plates or flasks.
Representative Results
Single cells were distinguished from debris and doublets on the basis of forward and side scatter distributions. Live cells were identified by their failure to take up the DAPI dye. The gating strategy was chosen on the basis of isotype control labelling of this live, whole-cardiac cell dissociation (Figure 1). From the live cells, CD45+ cells were first gated out, followed by CD56+ cells. CD144+ endothelial cells were then removed from the CD56– fraction. From this final population, CD146+/CD34– pericytes and CD146–/CD34+ adventitial cells were selected and subsequently collected (Figure 2). Immediately after cell collection, a small sample of each subpopulation (500-1,000 cells) was re-run through the sorter to confirm that cell distribution was as recorded in the initial sort. FACS-purified myocardial pericytes and adventitial cells both exhibit a spindle to stellate cell morphology in culture (Figure 3).
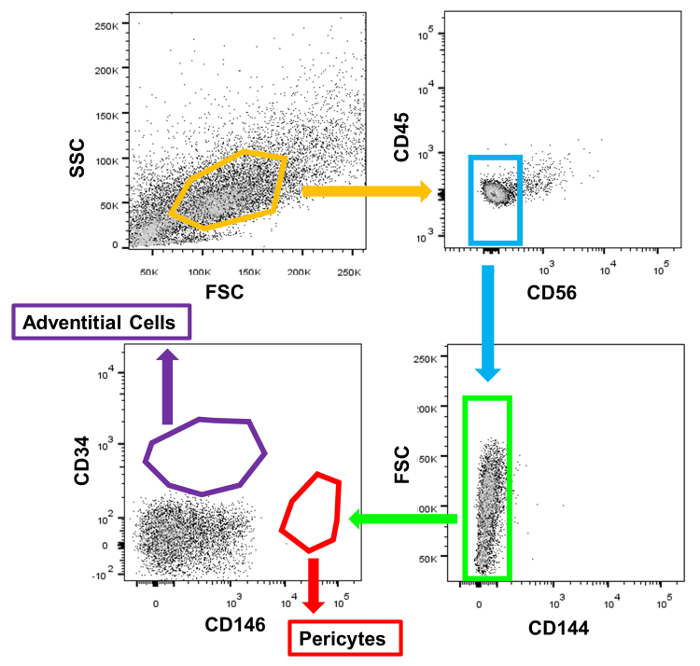
Figure 1: FACS Gating Strategy. Representative dots plots of isotype control labelled dissociated human heart cells illustrate the positioning of the sort gates.. Please click here to view a larger version of this figure.
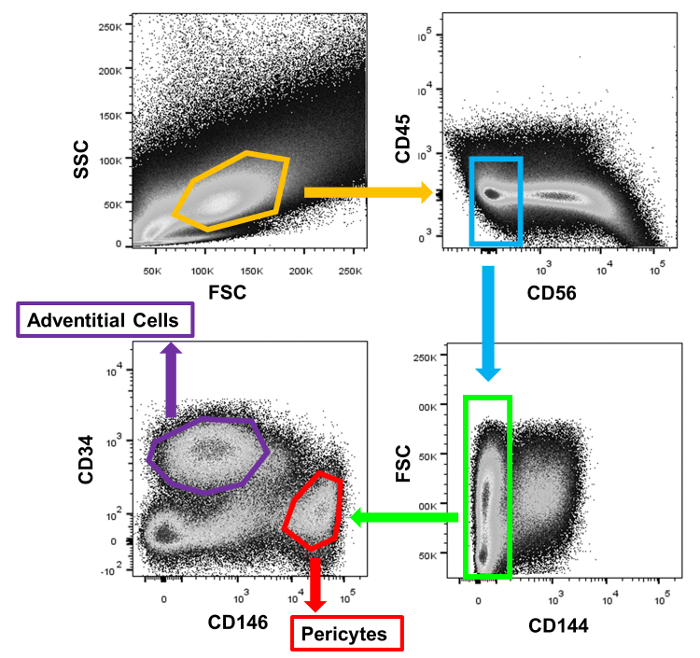
Figure 2: FACS Purification of Myocardial Perivascular Cells. Representative sorting of the two subpopulations of cardiac perivascular precursor cells from a single human myocardial sample. Please click here to view a larger version of this figure.
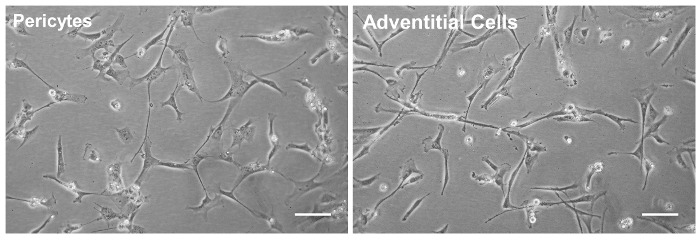
Figure 3: Perivascular Cell Morphology in Culture. The two cardiac perivascular precursor cell populations, pericytes (left panel) and adventitial cells (right) were sorted to homogeneity by FACS purification and further expanded in culture (at passage 3, Scale bars = 50 μm). Please click here to view a larger version of this figure.
Discussion
Increasing evidence supports a limited regenerative capacity of the adult human heart after injury. Identification and characterization of native precursor cells responsible for such regenerative responses in injured hearts are critical for both the understanding of associated mechanisms and signalling pathways and the development of approaches to utilize these cells therapeutically.
Previous protocols have described the isolation of perivascular precursor cell subsets from human skeletal muscle25. However, the direct application of these techniques to cardiac tissues often results in very poor cell yields. Consequently, we have made major adjustments to the protocol in order to enrich two discrete subpopulations of perivascular precursor cells, namely pericytes and adventitial cells, from human myocardial biopsies and to facilitate the simultaneous purification of both subsets. Isolation of both cell populations from the same sample not only optimizes use of precious cardiac tissue biopsies but also allows direct comparison of distinct perivascular cell subsets.
In order to obtain the maximum yield of cells, the freshness of the tissue sample is of critical importance. Maximal viable cell yields for pericytes and adventitial cells are approximately 3% and 4% of the total live cell dissociation respectively. Yields may vary greatly from around 30,000 to 400,000 cells per subset and are highly dependent on a number of factors, including the amount and quality of the starting tissue and the preservation duration. Perivascular cells are relatively delicate; samples showing evidence of autolytic change invariably yield low cell numbers after FACS. Similarly, perivascular cells are sensitive to harsh digestion techniques, and over-digestion will also result in a reduced viable cell yield. Customized adjustments of the digestion time and agitation speed may be necessary by individual laboratories. Finally, donor age and sample size will also have major impacts on cell yields.
We have extensively characterized cardiac pericytes obtained in this manner3. These cells demonstrated homogeneity in culture with no endothelial cell contamination. Population doubling times of around 60 hr between passages 3 and 12 were noted with the consistent expression of both common pericyte markers (alkaline phosphatase, NG2 and α-SMA) and classic MSC markers (CD44, CD73, CD90 and CD105) over this period. Intriguingly, it was found that after demethylation, a fraction of cardiac pericytes expressed the cardiomyogenic transcription factors Nkx2.5 and GATA4. When co-cultured with neonatal rat cardiomyocytes, a fraction of demethylated cardiac pericytes expressed the sarcomeric protein markers alpha-actinin and cardiac troponin-T, with a minor group further exhibiting spontaneous cytoplasmic calcium fluxes. Taken together, these findings suggest that a subpopulation of cardiac pericytes possesses cardiomyogenic potential and therefore may play a role in the intrinsic myocardial repair process. In comparison with pericytes, cardiac adventitial cells remain largely uncharacterized. Our unpublished preliminary data suggest that cardiac adventitial cells express some pericyte markers such as PDGFR-β and NG2, albeit at lower levels than cardiac pericytes, whilst losing CD34 in culture. These cells also express classic MSC markers and show osteogenic and adipogenic differentiation potential. Further studies of cardiac adventitial cells including angiogenic and pro-cardiogenic properties are ongoing.
The protocol described here enables simultaneous, prospective purification of cardiac pericytes and adventitial cells from a single human cardiac tissue sample. These cells represent novel cardiac precursor cell populations with potential cardiomyogenic properties that may prove to be suitable candidates for future cell therapies.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors wish to thank Shonna Johnston, Claire Cryer, Fiona Rossi and Will Ramsay at the University of Edinburgh and Alison Logar and Megan Blanchard at the University of Pittsburgh for their expert assistance with flow cytometry. We also wish to thank Anne Saunderson and Lindsay Mock for their help with obtaining human tissues. Human adult and fetal heart tissue samples were procured with full ethics permission of the NHS Scotland Tayside Committee on Medical Research Ethics and the NHS Lothian Research Ethics Committee (REC08/S1101/1) respectively. This work was supported by grants from the Medical Research Council (BP), British Heart Foundation (BP), Commonwealth of Pennsylvania (BP), Children’s Hospital of Pittsburgh (BP), National Institute of Health R01AR49684 (JH) and R21HL083057 (BP), and the Henry J. Mankin Endowed Chair at University of Pittsburgh (JH). JEB was supported by a British Heart Foundation Centre of Research Excellence doctoral training award (RE/08/001/23904). WC was supported in part by an American Heart Association predoctoral fellowship (11PRE7490001).
Materials
| AbC Anti-mouse Bead Kit | Molecular Probes | A-10344 | |
| Collagenase I | Gibco | 17100-017 | Reconstitute powder as required and filter sterilise |
| Collagenase II | Gibco | 17101-015 | |
| Collagenase IV | Gibco | 17104-019 | |
| anti-human CD34-PE | BD Pharmingen | 555822 | Keep sterile |
| anti-human CD45-APC-Cy7 | BD Pharmingen | 557833 | Keep sterile |
| anti-human CD56-PE-Cy7 | BD Pharmingen | 557747 | Keep sterile |
| anti-human CD144-PerCP-Cy5.5 | BD Pharmingen | 561566 | Keep sterile |
| anti-human CD146-AF647 | AbD Serotec | MCA2141A647 | Keep sterile |
| EGM2-BulletKit | Lonza | CC-3162 | For collection of cells and culture until adhered |
| DMEM, high glucose, GlutaMAX without sodium pyruvate | ThermoFischer Scientific | 10566-016 | |
| Fetal Bovine Serum | ThermoFischer Scientific | 10500-064 | Freeze in aliquots and keep sterile |
| Gelatin | Sigma Aldrich | G1393 | Dilute with sterile water |
| IgG1k-PE | BD Pharmingen | 559320 | Keep sterile |
| IgG1k-APC-Cy7 | BD Pharmingen | 557873 | Keep sterile |
| IgG1k-PE-Cy7 | BD Pharmingen | 557872 | Keep sterile |
| IgG1k-PerCP-Cy5.5 | BD Pharmingen | 561566 | Keep sterile |
| IgG1k-647 | AbD Serotec | MCA1209A647 | Keep sterile |
| Mouse serum | Sigma Aldrich | M5905 | Keep sterile |
| Paraffin Film – Parafilm M | Sigma Aldrich | P7793 | |
| Penicillin-Streptomycin | Gibco | 15979-063 | Freeze in aliquots and keep sterile |
| Phosphate buffered saline pH 7.4 | ThermoFischer Scientific | 10010-023 | Keep sterile |
| Red Blood Cell Lysing Buffer Hybri-Max | Sigma Aldrich | R7757 | Keep sterile |
| Trypan Blue Solution | Sigma Aldrich | T8154 | |
| Trypsin-EDTA 0.5%(10X) | Invitrogen | 15400-054 | |
| FACSARIA FUSION | BD Pharmingen | Fluorescence Activated Cell Sorter |
References
- Bergmann, O., et al. Evidence for cardiomyocyte renewal in humans. Science (New York, N.Y.). 324 (5923), 98-102 (2009).
- Laflamme, A., Murry, C. E. Heart regeneration. Nature. 473 (7347), 326-335 (2011).
- Chen, W. C. W., et al. Human myocardial pericytes: multipotent mesodermal precursors exhibiting cardiac specificity. Stem cells (Dayton, Ohio). 33 (2), 557-573 (2015).
- Campagnoli, C., Roberts, I. A., Kumar, S., Bennett, P. R., Bellantuono, I., Fisk, N. M. Identification of mesenchymal stem/progenitor cells in human first-trimester fetal. Blood. 98 (8), 2396-2402 (2001).
- Zuk, P. A., et al. Human adipose tissue is a source of multipotent stem cells. Molecular biology of the cell. 13 (12), 4279-4295 (2002).
- Chen, S., et al. Effect on left ventricular function of intracoronary transplantation of autologous bone marrow mesenchymal stem cell in patients with acute myocardial infarction. The American journal of cardiology. 94 (1), 92-95 (2004).
- Ringdén, O., et al. Mesenchymal stem cells for treatment of therapy-resistant graft-versus-host disease. Transplantation. 81 (10), 1390-1397 (2006).
- Kharaziha, P., et al. Improvement of liver function in liver cirrhosis patients after autologous mesenchymal stem cell injection: a phase I-II clinical trial. European journal of gastroenterology & hepatology. 21 (10), 1199-1205 (2009).
- Spaeth, E., Klopp, A., Dembinski, J., Andreeff, M., Marini, F. Inflammation and tumor microenvironments: defining the migratory itinerary of mesenchymal stem cells. Gene therapy. 15 (10), 730-738 (2008).
- Yan, X., et al. Injured microenvironment directly guides the differentiation of engrafted Flk-1(+) mesenchymal stem cell in lung. Experimental hematology. 35 (9), 1466-1475 (2007).
- Van Poll, D., et al. Mesenchymal stem cell-derived molecules directly modulate hepatocellular death and regeneration in vitro and in vivo. Hepatology (Baltimore, Md.). 47 (5), 1634-1643 (2008).
- Popp, F. C., et al. Mesenchymal stem cells can induce long-term acceptance of solid organ allografts in synergy with low-dose mycophenolate. Transplant immunology. 20 (1-2), 55-60 (2008).
- Li, Z., Zhang, C., Weiner, L. P., Zhang, Y., Zhong, J. F. Molecular characterization of heterogeneous mesenchymal stem cells with single-cell transcriptomes. Biotechnology advances. 31 (2), 312-317 (2013).
- Crisan, M., et al. A perivascular origin for mesenchymal stem cells in multiple human organs. Cell stem cell. 3 (3), 301-313 (2008).
- Ozerdem, U., Stallcup, W. B. Early contribution of pericytes to angiogenic sprouting and tube formation. Angiogenesis. 6 (3), 241-249 (2003).
- Rucker, H. K., Wynder, H. J., Thomas, W. E. Cellular mechanisms of CNS pericytes. Brain research bulletin. 51 (5), 363-369 (2000).
- Betsholtz, C. Insight into the physiological functions of PDGF through genetic studies in mice. Cytokine & Growth Factor Reviews. 15 (4), 215-228 (2004).
- Gerhardt, H., Betsholtz, C. Endothelial-pericyte interactions in angiogenesis. Cell and tissue research. 314 (1), 15-23 (2003).
- Crisan, M., Chen, C. W., Corselli, M., Andriolo, G., Lazzari, L., Péault, B. Perivascular multipotent progenitor cells in human organs. Annals of the New York Academy of Sciences. 1176, 118-123 (2009).
- Kang, S. G., et al. Isolation and perivascular localization of mesenchymal stem cells from mouse brain. Neurosurgery. 67 (3), 711-720 (2010).
- Chen, C. W., et al. Human pericytes for ischemic heart repair. Stem cells (Dayton, Ohio). 31 (2), 305-316 (2013).
- Campagnolo, P., et al. Human adult vena saphena contains perivascular progenitor cells endowed with clonogenic and proangiogenic potential. Circulation. 121 (15), 1735-1745 (2010).
- Katare, R., et al. Transplantation of human pericyte progenitor cells improves the repair of infarcted heart through activation of an angiogenic program involving micro-RNA-132. Circulation research. 109 (8), 894-906 (2011).
- Corselli, M., Chen, C. W., Sun, B., Yap, S., Rubin, J. P., Péault, B. The Tunica Adventitia of Human Arteries and Veins As a Source of Mesenchymal Stem Cells. Stem Cells and Development. 21 (8), 1299-1308 (2012).
- Crisan, M., et al. Purification and long-term culture of multipotent progenitor cells affiliated with the walls of human blood vessels: myoendothelial cells and pericytes. Methods in cell biology. 86, 295-309 (2008).

