Saccharomyces cerevisiae Metabolic Labeling with 4-thiouracil and the Quantification of Newly Synthesized mRNA As a Proxy for RNA Polymerase II Activity
Summary
The protocol described here is based on the genome-wide quantification of newly synthesized mRNA purified from yeast cells labeled with 4-thiouracil. This method allows to measure mRNA synthesis uncoupled from mRNA decay and, thus, provides an accurate measurement of RNA polymerase II transcription.
Abstract
Global defects in RNA polymerase II transcription might be overlooked by transcriptomic studies analyzing steady-state RNA. Indeed, the global decrease in mRNA synthesis has been shown to be compensated by a simultaneous decrease in mRNA degradation to restore normal steady-state levels. Hence, the genome-wide quantification of mRNA synthesis, independently from mRNA decay, is the best direct reflection of RNA polymerase II transcriptional activity. Here, we discuss a method using non-perturbing metabolic labeling of nascent RNAs in Saccharomyces cerevisiae (S. cerevisiae). Specifically, the cells are cultured for 6 min with a uracil analog, 4-thiouracil, and the labeled newly transcribed RNAs are purified and quantified to determine the synthesis rates of all individual mRNA. Moreover, using labeled Schizosaccharomyces pombe cells as internal standard allows comparing mRNA synthesis in different S. cerevisiae strains. Using this protocol and fitting the data with a dynamic kinetic model, the corresponding mRNA decay rates can be determined.
Introduction
Cells respond to endogenous and exogenous cues, through the dynamic alteration of their gene expression program. In recent years, a tremendous development of genome-wide methodologies allows the precise and comprehensive description of transcriptome changes in different conditions. In most transcriptomic studies, microarray hybridization or high-throughput sequencing are used to quantify RNA levels from a total steady-state RNA fraction. Transcriptional changes under a specific perturbation can display a wide range of possible outcomes, with either specific gene expression changes or a large spectrum of genes being either up- or downregulated. Gene expression is the result of a fine-tuned equilibrium — or steady-state — between RNA synthesis by RNA polymerases and other processes affecting RNA levels. RNA polymerase II transcription, including its three distinct phases (initiation, elongation, and termination), is highly and intricately associated with mRNA processing, cytoplasmic export, translation, and degradation.
Several recent studies demonstrated that mRNAs synthesis and decay are coupled mechanisms and showed that transcriptional effects upon mutation or under stimuli can be overlooked when quantifying total steady-state RNA. First, the detection of transcriptional changes through the analyses of steady-state levels of mRNA always depends on mRNAs half-life. Once the perturbation is introduced, the steady-state levels of mRNAs with long half-lives will be much less affected than those of mRNAs with short half-lives. Therefore, the detectability of the changes in RNA synthesis is strongly biased in favor of short-lived transcripts, while the analysis of longer-lived mRNA species might fail to reveal dynamic changes in transcription rate. Second, several reports have shown that, both in yeast and mammals, global changes in transcription might be overlooked when analyzing the steady-state levels of mRNA. This is likely due to the mechanisms that link mRNA synthesis and degradation resulting in mRNA buffering. This prompted the development of new protocols to quantify mRNA synthesis uncoupled from degradation, through the analysis of newly transcribed mRNA. In recent years, several alternatives have been presented, including global run-on sequencing (GRO-seq)1, and native elongation transcript sequencing (NET-seq)2,3. Here, we are presenting a protocol initially developed in mammalian cells4,5,6 and then adapted to yeast7,8,9,10,11, which is based on RNA labeling with a thiolated nucleoside or base analog, 4-thiouridine (4sU) or 4-thiouracil (4tU), respectively.
This method specifically purifies newly transcribed RNA from the cells in which RNA are pulse-labeled with 4sU with virtually no interference in the cell homeostasis. Hence, once the cells are exposed to 4sU, the molecule is rapidly uptaken, phosphorylated to 4sU-triphosphate, and incorporated in RNAs being transcribed. Once pulse-labeled, it is possible to extract total cellular RNA (corresponding to steady-state levels of RNA), and, subsequently, the 4sU-labeled RNA fraction is thiol-specifically modified, leading to the formation of a disulfide bond between biotin and the newly transcribed RNA4,5. However, 4sU can only be uptaken by the cells expressing a nucleoside transporter, like the human equilibrative nucleoside transporter (hENT1), preventing its immediate use in budding or fission yeast. While one could express hENT1 in either S. pombe or S. cerevisiae, an easier approach can be achieved using the modified base 4tU, since yeast cells can take up 4tU, without the need of expression of a nucleoside transporter10,11,12,13. In fact, the metabolism of 4tU requires the activity of the enzyme uracil phosphoribosyltransferase (UPRT). In several organisms, including yeast but not mammals, UPRT is essential for a pyrimidine salvage pathway, recycling uracil to uridine monophosphate.
An important bias in transcriptomic studies can be introduced by the normalization between different samples analyzed in parallel. Indeed, many deviating factors can affect the comparative analysis of the transcriptome of mutant and wild-type strains: the efficiency of cell lysis, differences in the extraction and recovery of RNA, and variances in scanner calibration for microarray analyses, among others. As discussed above, such variations can be particularly misleading when global effects on RNA polymerase II transcription are expected. An elegant mean to accurately compare mRNA synthesis rates between different samples was designed by using the distantly related fission yeast Schizosaccharomyces pombe as an internal standard. For that, a fixed number of labeled S. pombe cells is added to the S. cerevisiae samples, either wild-type or mutant cells, prior to cell lysis and RNA extraction10. Subsequently, both steady-state and newly synthesized RNAs from S. pombe and S. cerevisiae are quantified either by RT-qPCR or via the use of microarray chips or high-throughput sequencing10. Combining these data with kinetic modeling, absolute rates of mRNA synthesis and decay in budding yeast can be measured.
In the framework of this manuscript, we will show how the analysis of newly transcribed RNA allowed to reveal a global role for the coactivator complexes SAGA and TFIID in RNA polymerase II transcription in budding yeast14,15,16. Importantly, past studies quantified steady-state mRNA levels in S. cerevisiae and suggested that SAGA plays a predominant function on a limited set of yeast genes which are strongly affected by mutations in SAGA but relatively resistant to TFIID mutations17,18,19. Surprisingly, the SAGA enzymatic activities were shown to act on the whole transcribed genome, suggesting a broader role for this co-activator in RNA polymerase II transcription. Decreased RNA polymerase II recruitment at most expressed genes was observed upon the inactivation of SAGA or TFIID, suggesting that these coactivators work together at most genes. Hence, the quantification of newly transcribed mRNA revealed that SAGA and TFIID are required for the transcription of nearly all genes by RNA polymerase II14,15,16. The implementation of compensatory mechanisms emerges as a way for the cells to cope with a global decrease in mRNA synthesis which is buffered by a simultaneous global decrease in mRNA degradation. SAGA adds to the list of factors having a global effect on RNA polymerase II transcription, such as RNA Pol II subunits10, the Mediator coactivator complex20, the general transcription factor TFIIH21,22, and indirectly, elements of the mRNA degradation machinery9,10,23. Such compensatory events were universally observed in SAGA mutants, accounting for the modest and limited changes in steady-state mRNA levels despite a global and severe decrease in mRNA synthesis14. Similar analyses were also performed in a BRE1 deletion strain, resulting in a complete loss of histone H2B ubiquitination. Interestingly, a much milder but consistent global effect on RNA polymerase II transcription could be detected in the absence of Bre1, indicating that metabolic labeling of newly transcribed RNA in yeast can detect and quantify a wide range of changes in mRNA synthesis rates.
Protocol
1. Cell Culturing and Rapamycin Depletion of a SAGA Subunit
- For each S. cerevisiae strain and replicate, including wild-type or control strains, inoculate a single colony from a fresh plate onto 5 mL of YPD medium (2% Peptone, 1% yeast extract, and 2% glucose).
- Grow S. cerevisiae cells overnight at 30 °C with constant agitation (150 rpm).
- Measure the optical density at 600 nm (OD600) and dilute the culture to an OD600 of approximately 0.1 in 100 mL of YPD medium and let it grow up until the OD600 is around 0.8.
- In parallel, inoculate a single colony of S. pombe cells from a fresh plate onto 50 mL of YES medium (0.5% yeast extract; 250 mg/L adenine, histidine, uracil, leucine, and lysine; 3% glucose) and grow the cells overnight at 32 °C with constant agitation (150 rpm).
- Measure the OD600 of the S. pombe overnight culture and dilute the culture to an OD600 of approximately 0.1 in 500 mL of YES medium and let it grow up until the OD600 is approximately 0.8.
- For each S. cerevisiae anchor-away strain and replicate, including wild-type or control strains, inoculate a single colony from a fresh plate onto 5 mL of YPD medium (2% Peptone, 1% yeast extract, and 2% glucose).
- Grow the cells overnight at 30 °C with constant agitation (150 rpm).
- The next morning, measure the OD600, dilute the culture to an OD600 of approximately 0.1 in 100 mL of YPD medium and let it grow until the OD600≈ 0.8.
- Add 100 µL of rapamycin to the culture from a stock solution of 1 mg/mL (final rapamycin concentration of 1 µg/mL) and let the cells incubate at 30 °C with constant agitation for the time necessary for the protein of interest to be conditionally depleted from the nucleus (usually, 30 min is adequate). For the control, use a similar yeast culture, but instead of adding rapamycin, add the equivalent volume of dimethyl sulfoxide (DMSO).
2. 4tU Labeling with S. pombe as a Spike-in (Counting)
- Prepare a fresh solution of 2 M 4-thiouracil. Once prepared, keep it at room temperature and away from light.
- Accurately weigh 64.1 mg of 4-thiouracil for each S. cerevisiae culture and dissolve it in 250 µL of dimethylformamide (DMF) or in 250 µL of DMSO.
- For the S. pombe culture to be used as spike-in, weigh 320.5 mg of 4-thiouracil and dissolve it in 1,250 µL of DMSO.
- Add the 4-thiouracil solution to S. cerevisiae and S. pombe cultures for a final concentration of 5 mM and incubate them for 6 min with constant agitation at 30 °C and 32 °C, respectively.
- After 6 min, remove a small aliquot of each culture for cell counting. Count the cells using an automatic cell counter or a Neubauer chamber.
- Collect the cells through centrifugation (2,500 x g) for 5 min at 4 °C.
- Discard the supernatant, wash the cells with ice-cold 1x PBS, and centrifuge again (2,500 x g, 4 °C, 5 min).
- Calculate the total number of cells in each of the S. cerevisiae and S. pombe samples.
- Resuspend the cells in 5 mL of ice-cold 1x PBS and mix S. cerevisiae with S. pombe cells with a 3:1 ratio.
- Centrifuge the cells (2,500 x g, 4 °C, 5 min), remove the PBS, flash-freeze the cells in liquid N2, and store the sample at -80 °C until further use.
3. RNA Extraction and DNase Treatment
- Thaw the cells on ice for approximately 20–30 min.
- Proceed with the RNA extraction using a yeast-RNA extraction kit (Table of Materials) with a few adaptations.
- Per each sample, pour 750 µL of ice-cold Zirconia beads into a 1.5-mL screw cap tube supplied with the kit. Keep in mind that per each tube, RNA from up to 109 cells can be efficiently extracted. Hence, prepare the number of tubes necessary for each sample. For example, an S. cerevisiae culture of 100 mL (OD600≈ 0.8) can render around 2 x 109 to 3 x 109 cells, leading up to a total of 2.7 x 109 to 4 x 109 cells in total (after the spike-in with a third of the S. pombe cells). In this case, up to 3 – 4 reaction tubes per each sample/condition/mutant/replicate would be required.
- Per each 1 x 109 cells, add 480 µL of the lysis buffer provided with the kit, 48 µL of 10% SDS, and 480 µL of phenol:chloroform:isoamyl alcohol (25:24:1, v/v/v).
- Mix the cells using a vortex mixer and transfer them to the tubes containing the ice-cold Zirconia beads.
- Accommodate the tubes on a vortex mixer adaptor, turn the vortex at maximum speed, and beat for 10 min to lyse the yeast cells (in a room at 4 °C). Alternatively, perform lysis of the cells in an automatic bead-beater.
- Centrifuge tubes at 16,000 x g for 5 min at room temperature and carefully collect the upper phase (RNA-containing phase) to a fresh 15 mL falcon tube. Typically, the volume recovered per each tube is around 500–600 µL.
- To the 15 mL tubes containing the partially purified RNA, add the binding buffer provided with the kit and mix thoroughly. Per each 100 µL of RNA solution, add 350 µL of binding buffer (i.e., when the aqueous-phase solution volume is 600 µL, 2.1 mL of the binding buffer should be added).
- To the previous mixture, add the 100% ethanol and mix thoroughly. Per each 100 µL of RNA solution, add 235 µL of 100% ethanol (i.e., when the aqueous-phase solution volume is 600 µL, add 1.41 mL of ethanol).
- Apply up to 700 µL of the mixture from step 3.9 to a filter cartridge assembled in a collection tube, both provided with the kit.
- Centrifuge for 1 min at 16,000 x g. If the centrifugation duration was not enough for the total volume to pass through the filter, repeat the centrifugation for 30 s.
- Discard the flow-through and reuse the same collection tube. Add another 700 µL of RNA-binding buffer-ethanol solution to the filter and centrifuge again at 16,000 x g for 1 min.
- Discard the flow-through and repeat steps 3.11 and 3.12 until the RNA solution is finished.
- Wash the filter 2x with 700 µL of washing solution 1. Collect the washing solution via centrifugation at 16,000 x g for 1 min and always keep the collection tube.
- Wash the filter 2x with 500 µL of washing solution 2. Collect the washing solution via centrifugation at 16,000 x g for 1 min and always keep the collection tube.
- Centrifuge the tubes once again at 16,000 x g for 1 min to completely dry the filter.
- Transfer the filter cartridge to the final collection tube (RNA-appropriate tube) and elute RNA with 50 µL of DEPC-treated, RNase-free H2O (preheated to 100 °C).
- Centrifuge for 1 min at 16,000 x g.
- Elute RNA again (to the same tube) with 50 µL of preheated DEPC-treated, RNase-free H2O. Make sure that all the volume has passed through the filter; otherwise, centrifuge for longer periods.
- If multiple tubes for one single sample are used, pool them all in one tube.
- Quantify and check the purity of the sample using the appropriate equipment.
NOTE: While 4tU is only incorporated within newly synthesized RNA, there is a chance of minor contamination with DNA. For that reason, it is always advisable to treat the samples with DNase I. For that, use the reagents provided with the RNA-extraction kit (Table of Materials) following the manufacturer's recommendations.
4. Thiol-specific Biotinylation of Newly Synthesized RNA
- Adjust the concentration of the RNA obtained with section 3 of the protocol to 2 mg/mL. Aliquot 200 µg of total RNA, heat it for 10 min at 60 °C, and immediately chill it on ice for 2 min.
- To the RNA aliquot, add the reagents mentioned below in the following order: 600 µL of DEPC-treated, RNase-free H2O, 100 µL of biotinylation buffer (100 mM Tris-HCl [pH 7.5] and 10 mM EDTA, in DEPC-treated, RNase-free H2O), and 200 µL of biotin-HPDP from a stock of 1 mg/mL biotin-HPDP in DMSO or DMF.
- In some situations, the biotin-HPDP solution tends to precipitate, likely due to its low solubility in water. In this situation, increase the volume of DMSO/DMF up to 40% of the reaction volume (to the RNA sample, add 400 µL of DEPC-treated H2O, 100 µL of biotinylation buffer, and 400 µL of biotin-HPDP from a 0.5 mg/mL stock).
- Incubate the sample at room temperature and protected from light for 3 h, with gentle agitation.
- After incubation, add an approximately equal volume of chloroform to the tubes and mix vigorously.
- Spin the sample at 13,000 x g for 5 min, at 4 °C. This step allows the removal of excess biotin that did not biotinylate the RNA. Alternatively, perform this step using phase-lock tubes (heavy). For that, spin down the phase-lock tubes for 1 min at 13,000 x g, add the RNA mixture and an equal amount of chloroform, mix them vigorously, and centrifuge them at 13,000 x g for 5 min, at 4 °C.
- Carefully transfer the upper phase into new 2 mL tubes.
- Add one-tenth of the volume of 5 M NaCl and mix the sample.
- Add an equal volume of isopropanol, mix the sample thoroughly, and spin it at 13,000 x g for at least 30 min, at 4 °C.
- Cautiously remove the supernatant and add 1 mL of ice-cold 75% ethanol.
- Spin at 13,000 x g for 10 min, at 4 °C.
- Carefully remove supernatant, quick-spin the tube, and remove the remaining ethanol solution. Make sure that the RNA pellet does not dry.
- Suspend the RNA in 100 µL of DEPC-treated, RNase-free H2O.
5. Purification of Newly Synthesized Fraction from Total and Unlabeled RNA Using Streptavidin-coated Magnetic Beads
- Heat the biotinylated RNA for 10 min at 65 °C and then chill the samples on ice for 5 min.
- Add 100 µL of streptavidin-coated magnetic beads to the biotinylated RNA (at a final volume of 200 µL). Specifically, it is recommended to use the beads indicated in the Table of Materials, since, after conversations with other laboratories, these seemed to be the more consistent and reliable.
- Incubate the sample with slight shaking for 90 min, at room temperature.
- Place the columns provided with the kit (Table of Materials) in the magnetic stand.
- Add 900 µL of room-temperature washing buffer (100 mM Tris-HCl [pH 7.5], 10 mM EDTA, 1 M NaCl, and 0.1% Tween 20, in DEPC-treated, RNase-free H2O) to the columns (pre-run and equilibrate).
- Apply beads/RNA mixture (200 µL) to the columns.
- Collect the flow-through in 1.5-mL tubes and apply it again to the same magnetic column. If necessary, keep this flow-through as it represents the unlabeled RNA fraction.
- Wash the columns 5x with increasing volumes of washing buffer (600, 700, 800, 900, and 1,000 µL).
- Elute the newly synthesized RNA with 200 µL of 0.1 M DTT.
- Perform a second elution, 3 min later, with an equal volume of 0.1 M DTT.
- After eluting the RNA, add 0.1 volumes of 3 M NaOAc (pH 5.2), 3 volumes of ice-cold 100% ethanol, and 2 µL of 20 mg/mL glycogen (RNA-grade) and let the RNA precipitate overnight, at -20 °C.
- Recover the RNA by centrifugation (13,000 x g for 10 min, at 4 °C) and resuspend it in 15 µL of DEPC-treated, RNase-free H20. The proportion of labeled-to-total RNA is expected to be around 2% – 4% (usually more toward the lower end), rendering approximately 2.0 µg of newly synthesized RNA. This quantity is enough to do several qPCR experiments, as well for microarray/sequencing analyses.
6. RT-qPCR Validation of the Different Fractions
- Synthesize cDNA from 2 µg of total RNA or 10 µL of labeled RNA using random hexamers and the reverse transcriptase of choice, according to the manufacturer's instructions (Table of Materials).
- Amplify the cDNA by real-time qPCR using a standard protocol (Table of Materials).
NOTE: All samples should be run in triplicate from a minimum of two biological replicates. Correct all raw values for the expression of S. pombe tubulin.
7. Microarray Hybridization
- Hybridize RNA samples onto preferred microarray chips according to the manufacturer's instructions (for this specific protocol, see Table of Materials). Briefly, prepare biotinylated cRNA targets from 150 ng of RNA using the Premier RNA Amplification Kit (Table of Materials), according to the manufacturer's instructions. Hybridize 4 mg of fragmented cRNAs for 16 h at 45 °C and 60 rpm on microarray chips.
- Wash, stain, and scan the chips using the indicated station and scanner (Table of Materials). Extract the raw data (CEL Intensity files) from the scanned images using the command console (AGCC, version 4.1.2).
- Further process the CEL files with Expression Console software version 1.4.1 to calculate probe set signal intensities, using the statistics-based algorithms MAS 5.0 with default settings and global scaling as normalization method.
NOTE: The trimmed mean target intensity of each chip was arbitrarily set to 100. Perform all experiments using at least two independent biological replicates. Normalize raw data to the S. pombe signal and calculate fold changes in total and newly synthesized RNA levels.
8. Data Analysis Using an Existing R Pipeline
- Calculate synthesis and decay rates using a pipeline and R/Bioconductor package publicly available, as previously described8,10.
Representative Results
When performing metabolic labeling of newly transcribed RNA, several aspects need to be controlled: the time and efficiency of the labeling, the spike-in proportion, the extraction protocol, and the biotinylation efficacy (including signal-to-noise ratio), among others. These conditions have been extensively and methodically shown by others7,10,11. Here we mainly focus on the interpretation and immediate analyses that can be performed once the samples have been processed, either by RT-qPCR, microarray, or sequencing. The analyses of different mutant strains demonstrate the power of the method to detect not only a dramatic global decrease in mRNA synthesis, as in the case of SAGA mutant S. cerevisiae strains, but also a very mild reduction of RNA polymerase II activity upon suppression of histone H2B monoubiquitination.
Our analyses of SAGA enzymatic activities suggested a broad recruitment to chromatin15, which was not revealed by the analysis of steady-state mRNA levels in SAGA mutant strains. As RNA polymerase II recruitment was impaired upon SAGA inactivation, we decided to analyze whether mRNA synthesis rates would be globally affected. Hence, wild-type or mutant S. cerevisiae strains were exposed to 4tU for a 6 min period, to label newly transcribed RNAs. After mixing with the spike-in labeled cells (S. pombe) in a proportion of 3:1, total RNA was extracted, and newly synthesized RNA was biotinylated and purified according to the protocol presented here, as in the chronogram shown in Figure 1. Labeled RNAs were purified from a total of 200 µg of RNA, ensuring that the amount of purified product would be sufficient for any downstream application. As an initial and systematic step before any genome-wide analysis, newly synthesized RNA purification was validated by RT-qPCR. Genes were selected according to different parameters, including a level of expression, regulatory pathways, and a dependence on different RNA polymerases.
To confirm that this protocol specifically purifies labeled RNA, we quantified the levels of transcripts in fractions purified from wild-type cells that were cultured with or without 4tU. Negligible background levels of the analyzed RNAs were detected from cells that were not exposed to 4tU (Figure 2A). Newly transcribed RNA purification was further validated by the observed enrichment of the intron-containing ACT1 pre-mRNA (data not shown). After validation of the quality of the samples, we tested whether mRNA synthesis would be affected upon deletion of SPT20, which is known to disrupt the SAGA complex assembly. As reported by others, mRNA quantification performed on total (steady-state) RNA from the spt20Δ strain revealed mostly unchanged or mildly reduced levels for the tested genes (Figure 2B). Similar results were obtained for strains deleted for BRE1 (Figure 2B). In contrast, the analysis of newly transcribed RNA from the spt20Δ strain revealed a dramatic decrease in mRNA synthesis by three- to fivefold for all tested genes (Figure 2C). The loss of Bre1 led to a more discreet but still visible decrease in newly synthesized mRNA levels for the studied genes (Figure 2C). In good agreement with a role of SAGA and Bre1 in RNA polymerase II transcription, the loss of either Spt20 or Bre1 did not affect the expression of RDN25 or snR6 genes, transcribed by RNA polymerase I and RNA polymerase III, respectively (Figure 2C).
However, strains deleted for structural subunits of the SAGA complex, like SPT7 or SPT20, display severe slow-growth phenotypes which may account for the observed transcriptional alterations. To rule out undesired secondary effects, we conditionally depleted Spt7 from the nucleus, using anchor-away S. cerevisiae strains14,24. Upon 60 min of rapamycin treatment, prior to the pulse-labeling with 4tU (see Figure 1 for a schematic representation), newly transcribed mRNA levels were reduced to a similar extent as that observed in the deletion strain (Figure 2D and 2E). This analysis, thus, confirmed our former results and validated the protocol for this inducible depletion system. In a time-course analysis, where cells were exposed to rapamycin for a time spanning from 0 to 240 min, reduced expression was evidenced immediately after 15 min of exposure to the drug. More interestingly, steady-state mRNA levels tended to initially diminish but returned to normal levels after 60 min, an indication that a compensatory mechanism takes place in the meantime (Figure 2E).
Altogether, the labeling and quantification of newly synthesized RNA allowed revealing new regulatory roles for the SAGA complex. The described protocol could also reveal moderate effects on RNA polymerase II activity and was successfully applied to conditional depletion yeast strains.
One of the downstream applications for the purified newly synthesized RNA is a genome-wide quantification of transcripts using microarray hybridization or sequencing (4tU-seq). While high-throughput sequencing is more quantitative, sensitive and informative, microarray hybridization can be very helpful to determine whether the global mRNA levels are altered. In this context where normalization is pivotal, we added a spike-in organism to the sample that we aimed to analyze. Specifically, we mixed S. cerevisiae cells to S. pombe cells in a ratio of 3:1, both being previously exposed to 4tU. When the purified, labeled RNAs are subjected to high-throughput sequencing, any standard library preparation protocol can be used, most often following ribosomal RNA depletion. Normalization between 4tU-seq data from different samples has been performed by adding either labeled RNA from a different species (i.e., mixing S. cerevisiae and S. pombe cells as above)22 or in vitro-transcribed, thiolated spike-in RNA12,25,26,27. Commercially available microarray chips contain probes for the whole transcriptome of both budding and fission yeast, allowing the quantification of mRNA from both organisms in one single experiment. Microarray hybridizations were performed with total and newly synthesized RNA validated by RT-qPCR as indicated above (wild-type, spt20Δ, and bre1Δ). In addition, we included a strain that does not support monoubiquitination of histone H2B, through point mutation of the ubiquitinable residue (K123R). Because the exact same proportion of S. pombe cells were used in the different samples and replicates, it is possible to linearly rescale the arrays' intensities so that the total and labeled S. pombe probes have the same median intensity. This normalization, or rescaling, can be performed using a Bioconductor package developed by the laboratory of Patrick Cramer and is used in parallel in all analyzed samples, total and labeled fractions and wild-type and mutants strains8. Briefly, the input of this pipeline is an Excel file containing the probes and their intensity (MAS5 or RMA) for all the samples and fractions. After excluding probes that are outside the range of detection, the signal intensity is rescaled, taking into account the intensity values of the S. pombe probes. Finally, you can map the probes to the budding and fission yeast genomes, ending with a matrix containing the expression values normalized for the spike-in. These values can be further processed (see next section) or used as is. In the following example, we determined the fold change of every transcript between the mutant and the wild-type strain.
The analyses were performed for both steady-state or newly synthesized levels of RNA and plotted against their statistical significance (p-value) (Figure 3). In agreement with other studies, when total RNA levels were analyzed, only a few genes had their expression altered, either up- or downregulated (Figure 3A-3C). However, the analysis of newly transcribed RNA led to strikingly different conclusions. The newly transcribed mRNA levels of more than 4,000 genes were significantly reduced by at least twofold upon deletion of SPT20, suggesting a global positive effect of SAGA on RNA polymerase II transcription in budding yeasts (Figure 3D). Additionally, in the bre1Δ and K123R mutants, the results were more discreet: most genes appeared to have their expression reduced, but the extent of downregulation and the number of significantly affected genes (≈ 300 – 500) was indeed more limited (Figure 3E and 3F).
As previously mentioned, steady-state or total levels of mRNA are dictated by the tight equilibrium between synthesis and decay (Figure 4A). When RNA polymerase II transcription is globally impaired, two scenarios can be pictured: (i) either the total mRNA levels decrease globally, as a response to reduced synthesis but constant decay, or (ii) mRNA degradation is decreased to the same extent, resulting in mostly unchanged steady-state mRNA levels. The second scenario has been reported for several conditions, including in the context of SAGA or TFIID disruption9,10,14,16,22. A procedure called comparative dynamic transcriptome analysis (cDTA) based on 4tU labeling and dynamic kinetic modeling allows us to determine mRNA synthesis and infer the decay rates for every transcript8,10. Once again, we took advantage of the data collected for the previously mentioned strains (wild-type, spt20Δ, bre1Δ, and K123R). As expected, upon deletion of SPT20, we observed a simultaneous decrease in mRNA synthesis and degradation rates when compared to the wild-type strain. In this mutant strain, the compensation was almost optimal, with an average decrease in synthesis of 3.8-fold and an average decrease in decay of 4.1-fold (Figure 4B), corroborating why only limited transcriptional changes could be detected on total mRNA levels (Figure 3A). In the two other mutant strains (bre1Δ and K123R), concomitant changes in mRNA synthesis and decay were also observed, but the changes were on a much smaller scale and more dispersed (Figure 4C and 4D).
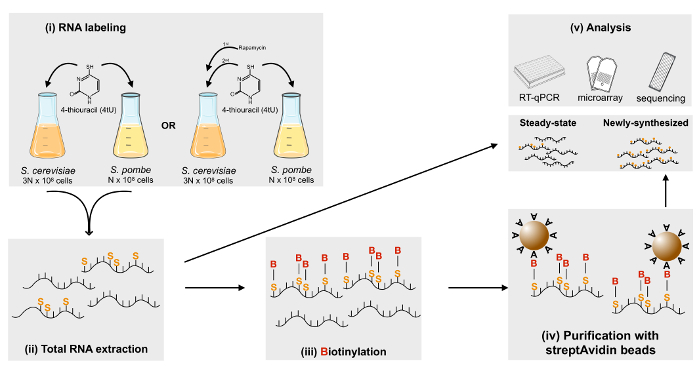
Figure 1: Schematic representation of the metabolic labeling of RNA using 4tU. Freshly prepared 4tU is added to the culture medium and cells are labeled for 6 min. Labeled S. cerevisiae and S. pombe cells are mixed in a ratio of 3:1 and total RNA is extracted. Afterward, newly synthesized RNA is biotinylated and can be purified using streptavidin-coated magnetic beads. Finally, total (steady-state) RNA and labeled newly synthesized RNA can be used in a variety of downstream applications, including RT-qPCR and microarray hybridization or sequencing. Please click here to view a larger version of this figure.
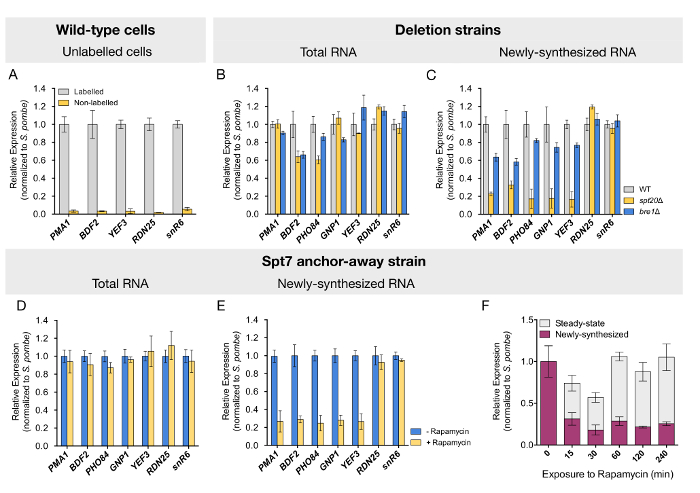
Figure 2: Analysis depicting transcriptional changes in both steady-state and newly transcribed RNA determined by RT-qPCR. (A) RNA levels of five different genes were quantified from the labeled RNA fraction purified from wild-type cells that were either exposed or not to 4tU. The next two panels show (B) total and (C) newly synthesized RNA quantification by RT-qPCR for wild-type (WT), spt20Δ, and bre1Δ yeast cells. Spt7 anchor-away strains, untreated or treated with rapamycin for 60 min, were labeled with 4tU, and (D) total or (E) newly transcribed RNA was quantified by RT-qPCR. (F) This panel shows the time-course analysis of changes in steady-state and newly synthesized mRNA upon Spt7 nuclear depletion. For all samples, mRNA levels for five RNA polymerase II genes were quantified by RT-qPCR. RNA polymerase I and RNA polymerase III genes (RDN25 and snR6, respectively) were used as control. Expression values (mean ± SD of three independent experiments) were normalized to the spiked-in S. pombe signal and set to 1 in the control sample. Panels D-F have been modified from Baptista et al.14. Please click here to view a larger version of this figure.
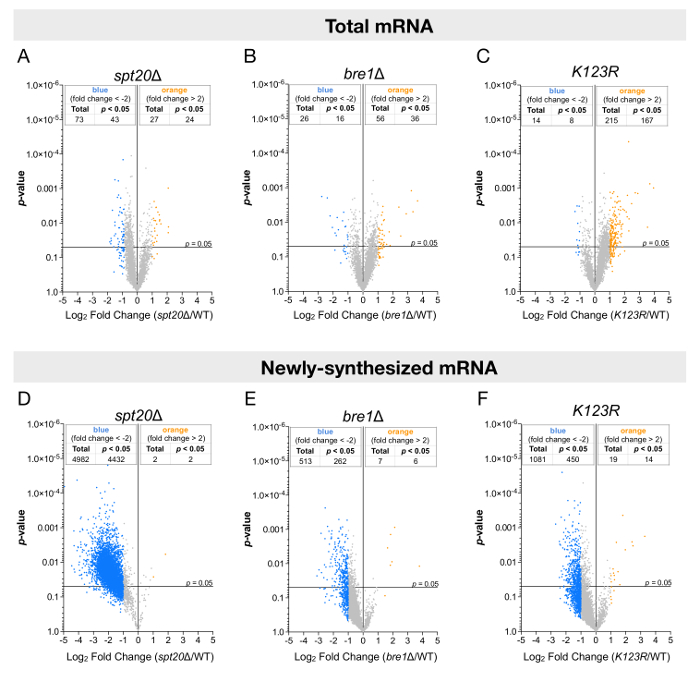
Figure 3: Genome-wide analyses of mRNA levels using total or labeled RNA fractions. These panels show volcano plots showing fold changes in (A-C) steady-state mRNA levels or (D-F) newly synthesized mRNA levels relative to their significance (p-value). The fold changes (FC) were calculated as the log2 of the ratio of the expression value of each gene after normalization to the S. pombe signal in the (A and D) spt20Δ, (B and E) bre1Δ, or (C and F) K123R strain versus the expression value of the same gene in wild-type S. cerevisiae. A total of 5,385 genes were analyzed, and thresholds of twofold change (blue dot: more than a twofold decrease; yellow dots: more than a twofold increase) and 0.05 p-values were considered. Panels A and D have been modified from Baptista et al.14. Please click here to view a larger version of this figure.
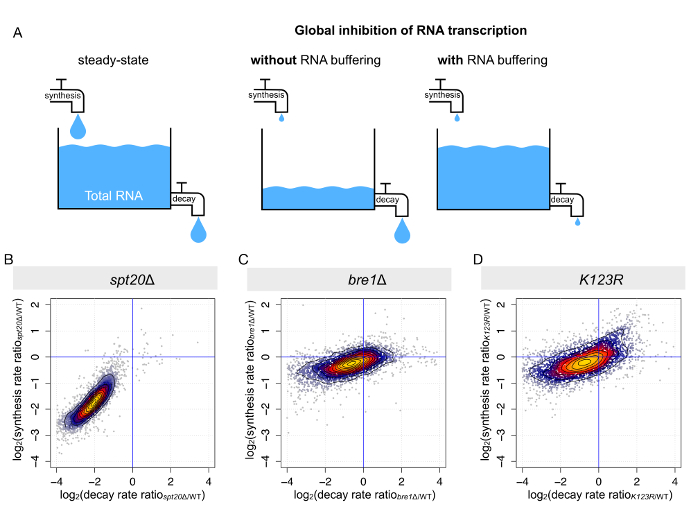
Figure 4: Parallel changes in mRNA synthesis and mRNA decay results in mRNA buffering. (A) This panel shows a schematic representation of the outcome of RNA synthesis perturbation on steady-state RNA levels. (B-D) These panels show the calculation of mRNA synthesis and the decay rates from the analyses of total and newly synthesized RNA. The synthesis and decay rates were determined for each S. cerevisiae transcript in (B) spt20Δ, (C) bre1Δ, and (D) K123R. Changes (calculated as the log2 of the ratio between mutant and wild-type) in synthesis rates were plotted against changes in decay rates. Please click here to view a larger version of this figure.
Discussion
While genome-wide tools to analyze changes in transcription are still improving, the sole analysis of the transcriptome through the quantification of steady-state levels of RNA might not accurately reflect changes in RNA polymerase II activity. Indeed, mRNA levels are regulated not only by RNA synthesis but also by their maturation and degradation. To measure mRNA synthesis uncoupled from mRNA degradation, distinct protocols have been developed in recent years for the analysis of nascent transcription in both yeast and mammals.
One of the most widely used protocols for the quantification of nascent transcription is GRO-seq1. While GRO-seq's main advantage is its capacity to unveil transcriptionally engaged and active polymerase at high resolution and low background, it has two distinct points that decrease its attractiveness: (i) it is technically challenging and requires the isolation of nuclei and, (ii) nuclei manipulations introduce some perturbations to the system28. Another alternative is NET-seq, which relies on the sequencing of the 3' ends of transcripts obtained upon immunoprecipitation of the extremely stable ternary complex formed between nascent RNA, template DNA, and RNA polymerase II. This approach allows the characterization of nascent RNAs at a base pair resolution and can explore mRNA processing events through immunoprecipitation of modified RNA polymerase II (serine-5 phosphorylation, for instance)2,3,29. Nevertheless, it presents some obstacles, from which we highlight three. First, while antibodies targeting RNA polymerase II are usually highly specific, it depends on antibody efficiency. Second, RNA might be prone to degradation during incubation periods, thus leading us back to the previous point (less efficient antibodies might require longer incubation times, leaving the RNA more susceptible to degradation)29. Third, and especially in mammals, the MNase digestion and size selection might exclude unique sequence alignment29,30.
Here we presented a detailed protocol for the metabolic labeling of newly synthesized RNA using 4tU in S. cerevisiae that has different advantages in comparison to other available approaches. Because transcription is highly sensitive to perturbations, cells should be maintained in the most physiological conditions. For instance, GRO-seq implies the arrest of transcriptionally engaged RNA polymerase II through exposure of the cells/nuclei to sarkosyl. However, sarkosyl treatment has been described as inhibitory of several cellular processes11. In this case, metabolic labeling with 4tU or 4sU at the described concentration is non-perturbing and does not visibly affect the cell homeostasis, especially for short periods of exposure. In contrast to other methods using transcription inhibition to measure mRNA half-lives, cDTA or 4tU-seq and fitting with dynamic modeling determines degradation rates of every single mRNA in unperturbed cells. Hence, one single method can simultaneously address both the synthesis and the decay rates of the whole transcriptome on a specific cell type. Since cDTA or 4tU-seq takes advantage of highly reliable normalization methods, namely through the usage of spike-in, different datasets can be analyzed together and directly compared. Finally, metabolic labeling and quantification of newly synthesized RNA is a technique that can be easily implemented in any molecular biology laboratory as it does not require any specific equipment. This likely explains the rather large dissemination of this technique, which tends to be used more and more systematically to explore RNA production and degradation in yeasts, as well as in higher eukaryotes.
RNA can be labeled in living cells using other nucleoside analogs, namely 5-bromouridine (BrU)31,32 or 5-ethynyluridine (EU)33,34. The isolation of BrU pulse-labeled RNA relies on anti-BrdU antibody purification which may have different efficiencies between experiments. EU-labeled RNA can be covalently conjugated to biotin using click chemistry leading to a non-reversible conjugation in contrast with thiol modification, which can be reversed with reducing agents. BrU and EU labeling have been used in mammalian cells to determine genome-wide RNA decay rates31,32 or to assess nascent RNA synthesis34,35. However, BrU or EU labeling has not been described in S. cerevisiae. Indeed, uptake of nucleoside analogs requires the expression of nucleoside transporter and, thus, these methods appear less flexible in budding yeast than labeling with 4tU.
The duration of 4tU labeling can be adapted and varies according to the question. When assessing mRNA synthesis in S. cerevisiae, a short labeling pulse of 6 min ensures that the newly transcribed mRNA levels are minimally affected by RNA degradation. Considering that the lag time before the modified nucleotide can be incorporated into the nascent RNA is less than 1 min, this 6-min labeling duration warrants that a reasonable amount of newly synthesized RNA can be purified. Such protocol, as defined by Miller et al.11, has been used in different S. cerevisiae mutants to reveal global changes in mRNA synthesis and RNA decay9,10,22,26,27,36. A longer labeling duration (3 h) was used in a metabolic pulse-chase labeling with 4tU to determine decay rates of all mRNAs in S. cerevisiae12. Finally, extremely short (from 1.5 min) 4tU labeling has been used to examine RNA processing kinetics in budding and fission yeast13,25,37.
Nevertheless, this method has some limitations, such as the relatively low yield of labeled RNA recovered at the end of the whole protocol. While in yeast, there is no limitation in the starting material, and this can be an issue when analyzing metazoan cells, specifically if this protocol is being utilized in vivo. One of the limiting steps of the protocol is biotinylation of the labeled RNA pool, whose efficiency is far from complete and was estimated to modify one in three 4sU residues in labeled RNA5. It recently showed that the use of methanethiosulfonate (MTS)-biotin, instead of biotin-HPDP, increases the yield of recovered RNA38. However, according to unpublished results from this research group and results from Rutkowski and Dölken, MTS-biotin is not fully thiol-specific, leading to the purification of unlabeled RNA, which is particularly problematic when low amounts of labeled RNA are purified39. Another limitation of the metabolic labeling using 4tU/4sU is the inherent speed at which RNA polymerase elongates. The average speed of RNA polymerase II was estimated to be approximately 3.5 kb/min40,41,42. Hence, in a 6 min labeling, the polymerase will have the capacity to transcribe 15–20 kb. Thus, in an experiment with a short pulse-labeling with 4tU/4sU, only the 3' end part of newly transcribed RNA is labeled, while the 5' end regions were pre-existing before the addition of 4tU/4sU. Thus, the purification of labeled RNAs also enriches pre-existing 5' ends of transcripts, particularly for long transcripts as in mammalian cells. In order to overcome this bias, in a refined protocol called TT-seq, the extracted labeled RNA is fragmented by sonication prior to the purification of newly synthesized species43.
Altogether, 4tU-seq is an excellent way to address transcriptional changes in a given context. Nevertheless, and while the protocol is fairly simple, it consists of several different steps, being ultimately relatively extensive. First and foremost, since this protocol handles RNA from start to finish, it is mandatory that all the requirements necessary for a clean and degradation-free sample are met. Hence, all reagents should be RNase-free, and all the materials should be dedicated to manipulating RNA, cleaning everything thoroughly and regularly with RNase decontamination solution. Second, while biotin reaction is highly specific, the excess of biotin-HPDP that did not react should be removed from the sample (to avoid the saturation of the streptavidin beads with biotin that is not covalently bound to RNA, for example). Third, as described in the protocol, the use of the indicated streptavidin-coated magnetic beads is highly recommended, since the several research groups we have talked to have all indicated that these beads led to lower background levels. Fourth, appropriate quality and experimental controls should be used. Include samples derived from cells that were not exposed to 4tU/4sU. These samples will be of great value to address background levels and to make sure that, during the experiment, a contamination with non-labeled RNA does not occur. Also, another excellent alternative to test whether the sample is enriched for newly synthesized RNA is to perform an RT-qPCR using primers against an intro-containing gene. The primers should be complementary to the 3' end and 5' end of two consecutive exon and intron, or vice versa. Finally, prior to the sequencing of microarray hybridization, validate the samples by RT-qPCR. For this, different genes with different levels of expression and regulation should be selected and analyzed. Optimally, this should be performed for the two different fractions of RNA (steady-state and newly synthesized RNA).
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Laszlo Tora for his support and V. Fisher, K. Schumacher, and F. El Saafin for their discussions. T.B. was supported by a Marie Curie-ITN fellowship (PITN-GA-2013-606806, NR-NET) and the Fondation ARC. This work was supported by funds from the Agence Nationale de la Recherche (ANR-15-CE11-0022 SAGA2). This study was also supported by ANR-10-LABX-0030-INRT, a French State fund managed by the Agence Nationale de la Recherche under the frame program Investissements d’Avenir ANR-10-IDEX-0002-02.
Materials
| 4-Thiouracil | Sigma-Aldrich | Cat# 440736 | |
| Rapamycin | Euromedex | Cat# SYN-1185 | |
| Countess II FL Automated Cell Counter | ThermoFisher | N/A | |
| RiboPure RNA Purification kit, yeast | ThermoFisher | Cat# AM1926 | |
| NanoDrop 2000 Spectrophotometer | ThermoFisher | ND-2000 | |
| TURBO DNA-free Kit | ThermoFisher | AM1907 | |
| EZ-Link HPDP Biotin | ThermoFisher | Cat# 21341 | |
| Thiolutin | Abcam | ab143556 | |
| µMACS Streptavidin kit | Miltenyi Biotec | Cat# 130-074-101 | |
| Transcriptor Reverse Transcriptase | Roche | 03 531 295 001 | |
| SYBR Green I Master | Roche | 4707516001 | |
| GeneChip Yeast Genome 2.0 | ThermoFisher | 900555 | |
| GeneChip Fluidics Station 450 | ThermoFisher | 00-0079 | |
| GeneChip Scanner 3000 7G | ThermoFisher | 00-0210 |
References
- Gardini, A. Global Run-On Sequencing (GRO-Seq). Methods in Molecular Biology. 1468, 111-120 (2017).
- Churchman, L. S., Weissman, J. S. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature. 469 (7330), 368-373 (2011).
- Churchman, L. S., Weissman, J. S. Native elongating transcript sequencing (NET-seq). Current Protocols in Molecular Biology. , 11-17 (2012).
- Cleary, M. D., Meiering, C. D., Jan, E., Guymon, R., Boothroyd, J. C. Biosynthetic labeling of RNA with uracil phosphoribosyltransferase allows cell-specific microarray analysis of mRNA synthesis and decay. Nature Biotechnology. 23 (2), 232-237 (2005).
- Dolken, L., et al. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 14 (9), 1959-1972 (2008).
- Kenzelmann, M., et al. Microarray analysis of newly synthesized RNA in cells and animals. Proceedings of the National Academy of Sciences of the United States of America. 104 (15), 6164-6169 (2007).
- Radle, B., et al. Metabolic labeling of newly transcribed RNA for high resolution gene expression profiling of RNA synthesis, processing and decay in cell culture. Journal of Visualized Experiments. (78), e50195 (2013).
- Schwalb, B., et al. Measurement of genome-wide RNA synthesis and decay rates with Dynamic Transcriptome Analysis (DTA). Bioinformatics. 28 (6), 884-885 (2012).
- Sun, M., et al. Global analysis of eukaryotic mRNA degradation reveals Xrn1-dependent buffering of transcript levels. Molecular Cell. 52 (1), 52-62 (2013).
- Sun, M., et al. Comparative dynamic transcriptome analysis (cDTA) reveals mutual feedback between mRNA synthesis and degradation. Genome Research. 22 (7), 1350-1359 (2012).
- Miller, C., et al. Dynamic transcriptome analysis measures rates of mRNA synthesis and decay in yeast. Molecular Systems Biology. 7, 458 (2011).
- Munchel, S. E., Shultzaberger, R. K., Takizawa, N., Weis, K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Molecular Biology of the Cell. 22 (15), 2787-2795 (2011).
- Eser, P., et al. Determinants of RNA metabolism in the Schizosaccharomyces pombe genome. Molecular Systems Biology. 12 (2), 857 (2016).
- Baptista, T., et al. SAGA Is a General Cofactor for RNA Polymerase II Transcription. Molecular Cell. 68 (1), 130-143 (2017).
- Bonnet, J., et al. The SAGA coactivator complex acts on the whole transcribed genome and is required for RNA polymerase II transcription. Genes & Development. 28 (18), 1999-2012 (2014).
- Warfield, L., et al. Transcription of Nearly All Yeast RNA Polymerase II-Transcribed Genes Is Dependent on Transcription Factor TFIID. Molecular Cell. 68 (1), 118-129 (2017).
- Huisinga, K. L., Pugh, B. F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Molecular Cell. 13 (4), 573-585 (2004).
- Lee, T. I., et al. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature. 405 (6787), 701-704 (2000).
- Lenstra, T. L., et al. The specificity and topology of chromatin interaction pathways in yeast. Molecular Cell. 42 (4), 536-549 (2011).
- Plaschka, C., et al. Architecture of the RNA polymerase II-Mediator core initiation complex. Nature. 518 (7539), 376-380 (2015).
- Helenius, K., et al. Requirement of TFIIH kinase subunit Mat1 for RNA Pol II C-terminal domain Ser5 phosphorylation, transcription and mRNA turnover. Nucleic Acids Research. 39 (12), 5025-5035 (2011).
- Rodriguez-Molina, J. B., Tseng, S. C., Simonett, S. P., Taunton, J., Ansari, A. Z. Engineered Covalent Inactivation of TFIIH-Kinase Reveals an Elongation Checkpoint and Results in Widespread mRNA Stabilization. Molecular Cell. 63 (3), 433-444 (2016).
- Haimovich, G., et al. Gene expression is circular: factors for mRNA degradation also foster mRNA synthesis. Cell. 153 (5), 1000-1011 (2013).
- Haruki, H., Nishikawa, J., Laemmli, U. K. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Molecular Cell. 31 (6), 925-932 (2008).
- Neymotin, B., Athanasiadou, R., Gresham, D. Determination of in vivo RNA kinetics using RATE-seq. RNA. 20 (10), 1645-1652 (2014).
- Shetty, A., et al. Spt5 Plays Vital Roles in the Control of Sense and Antisense Transcription Elongation. Molecular Cell. 66 (1), 77-88 (2017).
- Xu, Y., et al. Architecture of the RNA polymerase II-Paf1C-TFIIS transcription elongation complex. Nature Communications. 8, 15741 (2017).
- Adelman, K., Lis, J. T. Promoter-proximal pausing of RNA polymerase II: emerging roles in metazoans. Nature Reviews. Genetics. 13 (10), 720-731 (2012).
- Nojima, T., Gomes, T., Carmo-Fonseca, M., Proudfoot, N. J. Mammalian NET-seq analysis defines nascent RNA profiles and associated RNA processing genome-wide. Nature Protocols. 11 (3), 413-428 (2016).
- Storvall, H., Ramskold, D., Sandberg, R. Efficient and comprehensive representation of uniqueness for next-generation sequencing by minimum unique length analyses. PloS One. 8 (1), 53822 (2013).
- Tani, H., et al. Identification of hundreds of novel UPF1 target transcripts by direct determination of whole transcriptome stability. RNA Biology. 9 (11), 1370-1379 (2012).
- Tani, H., et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Research. 22 (5), 947-956 (2012).
- Jao, C. Y., Salic, A. Exploring RNA transcription and turnover in vivo. by using click chemistry. Proceedings of the National Academy of Sciences of the United States of America. 105 (41), 15779-15784 (2008).
- Palozola, K. C., et al. Mitotic transcription and waves of gene reactivation during mitotic exit. Science. 358 (6359), 119-122 (2017).
- Ardehali, M. B., et al. Polycomb Repressive Complex 2 Methylates Elongin A to Regulate Transcription. Molecular Cell. 68 (5), 872-884 (2017).
- Schulz, D., et al. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell. 155 (5), 1075-1087 (2013).
- Barrass, J. D., et al. Transcriptome-wide RNA processing kinetics revealed using extremely short 4tU labeling. Genome Biology. 16, 282 (2015).
- Duffy, E. E., et al. Tracking Distinct RNA Populations Using Efficient and Reversible Covalent Chemistry. Molecular Cell. 59 (5), 858-866 (2015).
- Rutkowski, A. J., Dolken, L. High-Resolution Gene Expression Profiling of RNA Synthesis, Processing, and Decay by Metabolic Labeling of Newly Transcribed RNA Using 4-Thiouridine. Methods in Molecular Biology. 1507, 129-140 (2017).
- Darzacq, X., et al. In vivo dynamics of RNA polymerase II transcription. Nature Structural & Molecular Biology. 14 (9), 796-806 (2007).
- Fuchs, G., et al. Simultaneous measurement of genome-wide transcription elongation speeds and rates of RNA polymerase II transition into active elongation with 4sUDRB-seq. Nature Protocols. 10 (4), 605-618 (2015).
- Singh, J., Padgett, R. A. Rates of in situ transcription and splicing in large human genes. Nature Structural & Molecular Biology. 16 (11), 1128-1133 (2009).
- Schwalb, B., et al. TT-seq maps the human transient transcriptome. Science. 352 (6290), 1225-1228 (2016).

