Efficient Differentiation of Human Pluripotent Stem Cells into Liver Cells
Summary
This protocol details a monolayer, serum-free method to efficiently generate hepatocyte-like cells from human pluripotent stem cells (hPSCs) in 18 days. This entails six steps as hPSCs sequentially differentiate into intermediate cell-types such as the primitive streak, definitive endoderm, posterior foregut and liver bud progenitors before forming hepatocyte-like cells.
Abstract
The liver detoxifies harmful substances, secretes vital proteins, and executes key metabolic activities, thus sustaining life. Consequently, liver failure—which can be caused by chronic alcohol intake, hepatitis, acute poisoning, or other insults—is a severe condition that can culminate in bleeding, jaundice, coma, and eventually death. However, approaches to treat liver failure, as well as studies of liver function and disease, have been stymied in part by the lack of a plentiful supply of human liver cells. To this end, this protocol details the efficient differentiation of human pluripotent stem cells (hPSCs) into hepatocyte-like cells, guided by a developmental roadmap that describes how liver fate is specified across six consecutive differentiation steps. By manipulating developmental signaling pathways to promote liver differentiation and to explicitly suppress the formation of unwanted cell fates, this method efficiently generates populations of human liver bud progenitors and hepatocyte-like cells by days 6 and 18 of PSC differentiation, respectively. This is achieved through the temporally-precise control of developmental signaling pathways, exerted by small molecules and growth factors in a serum-free culture medium. Differentiation in this system occurs in monolayers and yields hepatocyte-like cells that express characteristic hepatocyte enzymes and have the ability to engraft a mouse model of chronic liver failure. The ability to efficiently generate large numbers of human liver cells in vitro has ramifications for treatment of liver failure, for drug screening, and for mechanistic studies of liver disease.
Introduction
The purpose of this protocol is to efficiently differentiate human pluripotent stem cells (hPSCs) into enriched populations of liver bud progenitors and hepatocyte-like cells2. Access to a ready supply of human liver progenitors and hepatocyte-like cells will accelerate efforts to investigate liver function and disease and could enable new cellular transplantation therapies for liver failure3,4,5. This has proven challenging in the past since hPSCs (which include embryonic and induced pluripotent stem cells) can differentiate into all the cell-types of the human body; consequently, it has been difficult to exclusively differentiate them into a pure population of a single cell-type, such as liver cells6.
To precisely differentiate hPSCs into liver cells, first it is critical to understand not only how liver cells are specified but also how non-liver cell-types develop. Knowledge of how non-liver cells develop is important to logically suppress the formation of non-liver lineages during differentiation, thereby exclusively guiding hPSCs towards a liver fate2. Second, it is essential to delineate the multiple developmental steps through which hPSCs differentiate towards a liver fate. It is known that hPSCs sequentially differentiate into multiple cell-types known as the primitive streak (APS), definitive endoderm (DE), posterior foregut (PFG) and liver bud progenitors (LB) before forming hepatocyte-like cells (HEP). Earlier work revealed the signals specifying liver fate and the signals that suppressed the formation of alternate non-liver cell-types (including stomach, pancreatic, and intestinal progenitors) at each developmental lineage choice2,7,8.
Collectively, these insights have given rise to a serum-free, monolayer method to differentiate hPSCs towards primitive streak, definitive endoderm, posterior foregut, liver bud progenitors and finally, hepatocyte-like cells2. Overall the method involves the seeding of hPSCs in a monolayer at an appropriate density, preparing six cocktails of differentiation media (containing growth factors and small molecules that regulate various developmental signaling pathways), and sequentially adding these media to induce differentiation over the course of 18 days. During the process, no passaging of cells is needed. Of note, because this method explicitly includes signals that suppress the formation of non-liver cell-types, this differentiation approach1 more efficiently generates liver progenitors and hepatocyte-like cells by comparison to extant differentiation methods2,9,10,11,12. Furthermore, the protocol described in this text enables the faster generation of hepatocytes that ultimately express higher levels of hepatic transcription factors and enzymes than those produced by other protocols9,10,11,12.
The protocol described here has certain advantages over current differentiation protocols. First, it entails monolayer differentiation of hPSCs, which is technically simpler compared to three-dimensional differentiation methods, such as those that rely on embryoid bodies13. Second, this method exploits a recent advance whereby definitive endoderm cells (an early precursor to liver cells) can be efficiently and rapidly generated within 2 days of hPSC differentiation2,7, thus enabling the subsequent production of hepatocytes with increased purity. Third, in side-by-side comparisons, the hepatocyte-like cells produced by this method2 produce more ALBUMIN and express higher levels of hepatic transcription factors and enzymes compared to hepatocytes produced in other methods10,11,12.
Protocol
1. Preparation of Differentiation Media
NOTE: Refer to the Table of Materials for manufacturer information regarding the materials and reagents used.
- Preparation of base chemically defined media (CDM)
NOTE: CDM2, CDM3, CDM4 and CDM5 are chemically defined media that are used as base medias for differentiating hPSCs to liver cells at various stages. The composition of these medias can be found in Table 1.- To make CDM2 or CDM3, prepare a stock solution containing polyvinyl alcohol (PVA). Dissolve 0.5 g of PVA powder in 50 mL of Iscove’s Modified Dulbecco’s Medium (IMDM) to generate a 10 mg/mL PVA stock.
NOTE: Since PVA does not dissolve easily, prepare the PVA/IMDM mixture in a conical flask with continuous stirring at ~50 °C on a heating pad; stirring can be easily achieved using a magnetic stirrer. - After PVA is homogeneously dissolved in IMDM, remove the PVA solution from the heating pad and allow it to cool down to room temperature.
- Use a sterile, 0.2 µm filter to filter the PVA solution.
- Prepare the CDM2 and CDM3 by combining the filtered PVA from step 1.1.1 and various commercially bought components as outlined in Table 1 and the Table of Materials. Use sterile, 0.2 µm filter units to filter all media.
NOTE: Base medium can be stored at 4 °C but for no longer than 2 months. - Prepare the remaining base media CDM4 and CDM5 following Table 1.
- To make CDM2 or CDM3, prepare a stock solution containing polyvinyl alcohol (PVA). Dissolve 0.5 g of PVA powder in 50 mL of Iscove’s Modified Dulbecco’s Medium (IMDM) to generate a 10 mg/mL PVA stock.
- Preparation of differentiation media
- To prepare stock solutions for the small molecules and growth factors, reconstitute them as per the manufacturers’ recommendations. For storage, aliquot into sterile tubes to reduce freeze-thaw cycles and keep at -20 °C or as recommended.
NOTE: Composition of differentiation medium to be used at various differentiation stages is described in Table 2 and in the following steps. - To prepare the differentiation medium, first thaw the frozen small molecules and/or growth factors at room temperature. Next, aliquot out the required amount of base medium. Last, prepare the final differentiation media by adding the specified small molecules and growth factors to the base medium at the appropriate concentrations (Table 2).
NOTE: Freshly-prepared differentiation medium should ideally be used on the same day; otherwise it can be stored at 4 °C and used within three days. - Using a pipette tip, mix the differentiation medium several times to ensure supplements are homogeneously distributed before adding the medium to cells (e.g., add 1 mL of differentiation medium to each well of a 12-well plate.)
- To prepare stock solutions for the small molecules and growth factors, reconstitute them as per the manufacturers’ recommendations. For storage, aliquot into sterile tubes to reduce freeze-thaw cycles and keep at -20 °C or as recommended.
2. Seed hPSCs onto Plates at Defined Densities for Differentiation
- Coat cell culture plastics that will be used for seeding with hPSCs.
- Thaw the matrix (e.g., Geltrex) at 4 °C overnight before the day of use.
- The next day, dilute the matrix 1:100 by adding 500 μL of the matrix into 50 mL of cold Dulbecco’s Modified Eagle’s Medium (DMEM)/F12. Since the matrix is a hydrogel that irreversibly polymerizes upon exposure to room temperature, when working with the matrix, always keep the matrix tubes and media on ice.
- NOTE: The matrix dissolved 1:100 in DMEM/F12 can be stored at 4 °C but should be used within 2 months.
- To coat a culture plate with the matrix, pipette the diluted matrix into the required number of wells using just enough volume of matrix solution to cover the surface of the well (e.g., add 0.5 mL of matrix to one well of a 12-well plate or 1 mL to one well of a 6-well plate). If needed, shake the plate gently to make sure that the matrix solution has fully covered the bottom of the well.
- Leave the matrix-coated plate in a 37 °C incubator for at least 60 min. At this temperature, the matrix polymerizes to form a thin film at the bottom of the well.
- NOTE: Matrix-coated plates can be kept in the 37 °C incubator and used within 3 days as long as the matrix has not dried up.
- Aspirate the remaining matrix solution from the coated wells immediately prior to seeding the wells with hPSCs.
- Passaging and seeding the coated plates with hPSCsNOTE: The dissociation agent contains enzymes that dissociate cells and it is important to leave hPSCs in the dissociation agent for as short of a timespan as possible.
- To seed matrix-coated culture plates with hPSCs prior to differentiation, grow undifferentiated hPSCs to >70% confluency in commercially-available mTeSR1 medium (Table of Materials) according to the manufacturer’s protocol. It is critical to passage hPSCs before they become fully confluent, as hPSCs may spontaneously differentiate at high confluence.
NOTE: hPSCs must be karyotyped to ensure that there are no chromosomal abnormalities before using them for experiments. The hPSCs used for this differentiation protocol were maintained in mTeSR1 media and passaged as clumps with EDTA to minimize karyotypic abnormalities. Karyotypically-abnormal hPSCs should not be used for differentiation, as they may proliferate abnormally quickly; thus the cell-seeding densities recommended here are not appropriate for karyotypically-abnormal cells. - To seed hPSCs for differentiation, aspirate mTeSR1 from largely-confluent hPSC cultures plate and add commercially bought dissociation agent (Table of Materials) to dissociate the hPSCs, using just enough dissociation agent to cover the surface of the well or dish on which the cells are growing (e.g., add 0.5 mL of the dissociation agent per well of a 12-well plate, 1 mL per well of a 6-well plate or 3 mL per 10 cm dish).
- Incubate hPSCs in the dissociation agent at 37 °C for 5 min or until some colonies begin detaching. Gently tap the bottom of the well/plate several times; after several minutes of dissociation, most hPSC colonies should freely come into suspension.
- To dislodge dissociated hPSCs from the plate, add 2 mL/well of DMEM/F12 if working with a 6-well plate (or 1 mL/well if working with a 12-well plate) to dilute the dissociation agent. Use a 5 mL serological pipette to gently pipette up and down multiple times to wash off all cells from the surface of the well; collect resuspended single cells in a 50 mL conical tube. Wash the plate a second time with the same volume of DMEM/F12 to ensure recovery of all hPSCs.
- To the 50 mL conical tube containing the cells, add DMEM/F12 to dilute the original volume of the dissociation agent by 1:5-1:10 (e.g., if the original volume of the dissociation agent was 1 mL, adjust the total volume of cell suspension to 10 mL with DMEM/F12 to dilute the dissociation agent at 1:10)
- Centrifuge the collected hPSCs in a 50 mL conical tube at 350 x g for 3 min at 4 °C to pellet cells.
- While waiting for the cells to pellet, aspirate matrix from the plate in which the hPSCS will be seeded. Next, add sufficient amounts of mTesR1 supplemented with 1 μM of commercially-obtained thiazovivin (a pharmacological ROCK inhibitor) to recipient wells to cover them (e.g., add 0.5 mL mTeSR1 per well of a 12-well plate or 1 mL of mTeSR1 per well of a 6-well plate).
NOTE: Thiazovivin at a low concentration is included at this step to enhance single cell survival and hence the subsequent seeding density of hPSCs. - After centrifuging hPSCs, carefully aspirate the supernatant, leaving the pelleted hPSCs at the bottom of the conical tube. The supernatant contains dissociation agent which will inhibit subsequent adhesion of hPSCs and thus, it is important to aspirate the large majority of the supernatant before proceeding.
- Resuspend the cell pellet in mTeSR1 supplemented with 1 μM of thiazovivin. With a p1000 pipette, gently triturate 2-3 times to evenly resuspend the cell pellet into a single cell suspension. Do not over-triturate the cell pellet as excessive mechanical force will damage hPSCs and lead to poor cell survival.
- After resuspension of the hPSCs, immediately pipette 10 μL of the suspension into a hemocytometer and count the number of cells. It is important to pipette cells for counting as quickly as possible, as gravity will lead to hPSCs naturally settling into the bottom of the 50 mL tube, which will confound accurate cell counting.
- Adjust the volume of the resuspended hPSCs with thiazovivin-supplemented mTeSR1 to achieve the desired cell concentration for plating. For example, after adjusting the volume of resuspended hPSCs, add 0.5 mL of the cell suspension per well to seed 160,000-250,000 cells into each well of a 12-well plate, thus achieving a total volume of 1 mL in each well (0.5 mL of media was added to the well in step 2.2.7). If larger or smaller wells are being used, scale cell numbers/media volumes up or down accordingly.
NOTE: It is important to seed hPSCs at the indicated cell density. Overly-confluent hPSCs will not differentiate efficiently and under-confluent hPSCs will not survive well during differentiation. - Shake the plate in a cross pattern (left, then right; forward, then backward) several times to make sure cells are evenly distributed across the plate/well. Do not swirl the plate in a circular motion, as the cells will settle in the center of the plate/well. Typically, hPSCs will begin adhering to the surface of the well within ~3-30 min. Allow cells to grow for at least 24 h before initiating differentiation.
- To seed matrix-coated culture plates with hPSCs prior to differentiation, grow undifferentiated hPSCs to >70% confluency in commercially-available mTeSR1 medium (Table of Materials) according to the manufacturer’s protocol. It is critical to passage hPSCs before they become fully confluent, as hPSCs may spontaneously differentiate at high confluence.
3. Differentiation of hPSCs into Endodermal Cells and Liver Progenitors
- After hPSCs have been plated for at least 24 hours (as described in step 2.2.12), before proceeding with differentiation, check the morphology of cells under a phase-contrast microscope, with specific emphasis on the diameter and density of plated hPSC colonies.
NOTE: Ideally, clumps should be readily spaced and of appropriate size and density throughout the well before step 3.2. Large (approximately >400 μm) or small colonies (approximately <200 μm) of hPSCs are not usable for differentiation (Figure 4). Differentiation signals will not act evenly throughout large hPSC colonies, leading to inefficient differentiation. Colonies that are too small do not survive well during the first 3 days of hPSC differentiation in this differentiation protocol. If the density of seeded hPSCs is correct, the hPSC colonies will often form “bridges” of cells between them and the overall confluence will be ~30-50% before adding day 1 differentiation medium (Figure 4). - If colony sizes are ideal in step 3.1, proceed to day 1 of differentiation, which entails differentiation of hPSCs into anterior primitive streak (APS).
- Prepare day 1 APS differentiation medium by mixing all reagents outlined in Table 2 using CDM 2 (Table 1 and section 1.2) as the base medium. Pipet to mix several times to ensure even distribution of the components in the media.
- Aspirate to remove the thiazovivin supplemented mTeSR1 from the plated hPSCs and briefly wash hPSCs with IMDM media. Do not wash with phosphate buffered saline (PBS) instead of IMDM, as PBS lacks calcium ions (Ca2+) and is thus toxic to hPSCs; it will disrupt cell morphology.
- After the brief IMDM wash, add day 1 medium to the hPSCs. Record the time and place cells back into the 37 °C incubator
- Continue with the subsequent differentiation steps by preparing (Table 2) and adding the respective differentiation medium to the cells on the respective days (at 24 h intervals) of differentiation around the same time of the day (Figure 1). Replace with fresh media daily, even if consecutive days of differentiation use the same differentiation media. Between media changes, wash cells once with IMDM media to remove dead cells and remnant of previous medium components.
- As differentiation progresses beyond the liver bud stage, cell numbers increase and hence, add more media to each well of the plate to ensure that the amount of differentiation factors and nutrients is not limiting. For example, add 1 mL differentiation medium to each well of 12-well initially on days 1 to 6 of differentiation but add 1.5-2 mL of subsequent differentiation media on later days of differentiation (days 7 to 18 of differentiation).
4. Characterization of Endodermal Cells and Liver Progenitors by Immunostaining
- Prepare blocking buffer: 10% donkey serum + 0.1% Triton X100 in deionized phosphate buffered saline (DPBS).
- Prepare staining buffer: 1% donkey serum + 0.1% Triton X100 in DPBS.
- Aspirate medium from cells in 12-well plate.
- Add 4% paraformaldehyde (in DPBS) for 15 min at room temperature to fix cells and then wash the cells twice with DPBS.
- Add blocking buffer for 1 h at room temperature to block and permeabilize the fixed cells.
- Aspirate the blocking solution and add primary antibody diluted in staining buffer. (See Table of Materials for dilution ratios of antibodies.)
- Stain the cells overnight at 4 °C.
- Wash cells thrice with 0.1% Triton X100 in DPBS.
- Add secondary antibody stain in the staining buffer for 1 h at room temperature. (See Table of Materials for dilutions of secondary antibodies.)
- Remove the secondary antibody and add DAPI for 5 min at room temperature to conduct nuclear counterstain.
- Wash twice with 0.1% Triton X100 in DPBS to remove excess antibody and DAPI.
- Conduct fluorescence microscopy with a Zeiss Observer D1. Alternatively, store plate in 4 °C until imaging is performed. For expected results, see Figure 3.
5. Characterization of Liver Progenitors by Fluorescence Activated Cell-sorting (FACS) Analysis
NOTE: Use FACS to precisely quantify the percentage of AFP+ differentiated LB cells that emerge by day 6 of differentiation. Follow the same steps to quantify the percentage of ALB+ differentiated hepatocytes by day 18 of differentiation.
- Conjugate an anti-AFP antibody.
- Add R-phycoerythrin to anti-AFP antibody at a concentration of (0.55 µg/µL) using R-phycoerythrin conjugation kit (see Table of Materials).
NOTE: Ensure that the antibodies are purified and are reconstituted in buffers that do not contain amines. Ensure that amount of antibody used in a labeling reaction must be less than the amount of PE (i.e., 60 µg of antibody with 100 µg of PE). Poor conjugation of antibodies may lead to unreliable outcomes. - Add 1 µL of modifier reagent for 10 µL of antibody to be labeled. Mix gently.
- Remove the R-PE mix (100 µL) and pipette the above mixture directly into the lyophilized R-PE material.
- Place the cap back and leave the vial standing for 3 h in the dark at room temperature.
- After incubating for 3 h or more, add 1 µL of quencher reagent for 10 µL of antibody used. The conjugate can be used after 30 min.
- Store conjugated antibody at 4 °C.
- Wash differentiated or undifferentiated hPSCs in 6-well format with DMEM/F12.
- Briefly treat with dissociation agent (1 mL/well in a 6-well plate) for 5 min at room temperature until cells detach. Harvest and stain both undifferentiated hPSC and differentiated liver progenitor cells identically and analyze in parallel in the same experiment to ensure specificity of antibody staining.
- Gently tap Petri dish to detach cells. Use a p1000 pipette to detach cells off the plate and collect cells in a 50 mL conical tube. Ensure that cells have mostly detached before proceeding with the next step. Subsequently, wash wells twice more with 1xDPBS buffer to collect residual cells.
- In the 50mL conical tube, dilute dissociation agent in ~5 volumes of 1x DPBS buffer. Triturate rigorously 3-4 times with a p1000 pipette to ensure all cells are dissociated into single cells.
NOTE: It is imperative to generate single cells prior to centrifugation or clumps subsequently cannot be dissociated with ease. However, do not over-triturate as this may damage cell integrity. Count the number of cells and use the recommended proportion of cells to antibody ratio, which has been previously optimized for minimal background and maximal signal detection. - Centrifuge conical tube containing the cell suspension at 300 x g for 5 min at 4 °C.
- Aspirate the supernatant with care not to disturb the cell pellet.
- Resuspend cell pellet thoroughly in fixation/perm buffer to generate a single-cell suspension and fix on ice at 4 °C for 20 min. Note to transfer to a 2 mL microcentrifuge tube. Furthermore, usage of a 2 mL microcentrifuge tube at this step enables the cell pellet to deposit at the V-shaped-corner of the tube, minimizing cell loss during washes.
- Wash each pellet with 1.8 mL of perm/wash buffer twice. Resuspend cell pellet thoroughly in perm/wash buffer by pipette mixing 6 times with a p1000 pipette. Then, centrifuge, remove supernatant and repeat the wash process with 1.8 mL of perm/wash buffer.
- Resuspend cell pellet in perm/wash buffer so that there will be 100 µL/individual stain and subsequently transfer the cell suspension into a 2 mL microcentrifuge tube.
NOTE: It is imperative to generate a single-cell suspension at this step prior to antibody staining. Aggregates of cells will not be stained and thus will confound FACS analysis. Furthermore, usage of a 2 mL microcentrifuge tube at this step enables the cell pellet to deposit at the V-shaped-corner of the tube, minimizing cell loss during washes. - After resuspending cells in FACS buffer, aliquot them into individual 2 mL tubes (for both unstained control and antibody-stained samples).
- Stain with anti-AFP-PE 0.33 µL per 150,000 cells for 30 min at room temperature in the dark. For example, for a 100 µL individual stain, stain as follows: 0.33 µL of a-AFP PE and 100 µL of perm/wash buffer. Resuspend cells with p200 pipette well to ensure even staining.
NOTE: Only pipette-mix and do not vortex as this may reduce protein stability. - Wash, resuspend the cells twice in 1-2 mL of perm/wash buffer (1.9 mL/individual stain) and centrifuge at 800 x g for 5 min at 4 °C. Wash cells with no less than 1-2 mL of perm/wash buffer to ensure sufficient washing.
NOTE: Only pipette mix and do not vortex as this may reduce protein stability. After centrifuging, aspirate supernatant carefully to prevent cells from dislodging and remove as much supernatant as possible to minimize antibody carry-over and ensure a more complete wash. - Resuspend each washed pellet in 300 μL of perm/wash buffer and strain it through a 100 μm filter into a FACS tube to strain out large clumps of cells prior to FACS analysis.
- Analyze cells on a FACS Aria flow cytometry on the PE channel. Analyze a minimum of 10,000 events for each individual stain, and parse events by virtue of FSC-A/SSC-A analysis, select cell singlets by gating on FSC-W/FSC-H followed by SSC-H/SSC-W.
Representative Results
After 24 h of APS differentiation, colonies will generally adopt a different morphology than undifferentiated colonies concomitant with a loss of the bright border that typically circumscribes hPSC colonies. Morphologically, primitive streak cells generally have ragged borders and are more spread and less compact than hPSCs-this is evocative of an epithelial-to-mesenchymal transition as pluripotent epiblast cells differentiate and ingress into the primitive streak in vivo. If the colony size of the hPSCs prior to differentiation was too large, there will be an overtly-raised center that comprises undifferentiated cells. Cultures containing such large clumps should be discarded, as these undifferentiated colony centers will confound subsequent differentiation. If clumps of the correct size and density were plated, APS differentiation is highly reproducible and uniform, generating a 99.3±0.1% MIXL1-Gfp+ primitive streak population (MIXL1 is a primitive streak marker)7. Note that some cell death is observed after 24 h of APS differentiation.
By day 2 of differentiation, primitive streak cells have differentiated into day 2 definitive endoderm cells, the vast majority of which express SOX17 (Figure 2) and FOXA2. In dozens of independent experiments with 14 hESC and hiPSC lines (including H1, H7, H9, HES2, HES3, BJC1, BJC3, HUF1C4 and HUF58C4), this approach scalably, consistently and efficiently generated pure definitive endoderm populations (94.0% ± 3.1%)7. This method to generate definitive endoderm from hPSCs yields higher percentages of CXCR4+PDGFRA– endodermal populations compared to other endoderm-forming approaches2,14.
By day 3 of differentiation, endoderm has differentiated into foregut progenitors that appear polygonal in shape (Figure 1). Later, by 6 days of differentiation, the foregut progenitors differentiate into liver bud progenitors expressing AFP, TBX3, and HNF4A (Figure 3). Across three hESC lines (H1, H7, H9 hESCs), this method generates day 6 AFP+ liver progenitors form at an efficiency of 89.0±3.1% (Figure 2). Finally, after 18 days of liver differentiation from hPSCs, ALBUMIN+ hepatocyte-like cells appear. Morphologically, they appear epithelial, forming bright borders reminiscent of bile canaliculi (Figure 1). At this stage, the cytoplasm of the day 18 hPSC-derived hepatocytes appears darker than the nucleus (Figure 1).
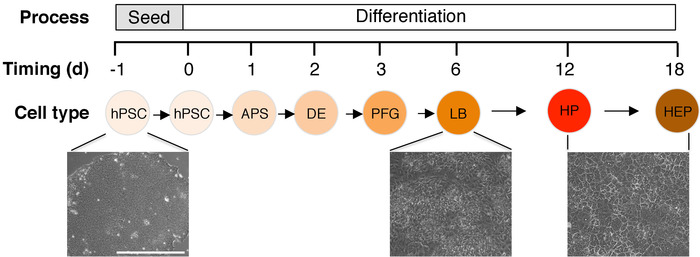
Figure 1: Schematic of differentiation strategy and morphology of undifferentiated hPSCs, differentiating endoderm and hepatocyte-like cells. Differentiation process and timeline were shown. Abbreviations: d = day, hPSC = human pluripotent stem cells, APS = anterior primitive streak, DE = definitive endoderm, PFG = posterior foregut, LB = liver bud progenitors, HP = hepatocyte progenitors, HEP = hepatocyte like cells. Scale bar = 400 μm. Please click here to view a larger version of this figure.
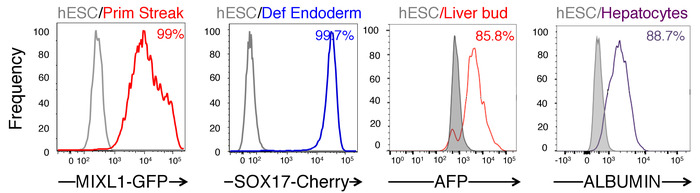
Figure 2: Percentage of day 1 MIXL1+ primitive streak, day 2 SOX17+ definitive (def) endoderm, day 6 AFP+ liver bud progenitors and ALB+ hepatocyte populations as shown by FACS. Please click here to view a larger version of this figure.
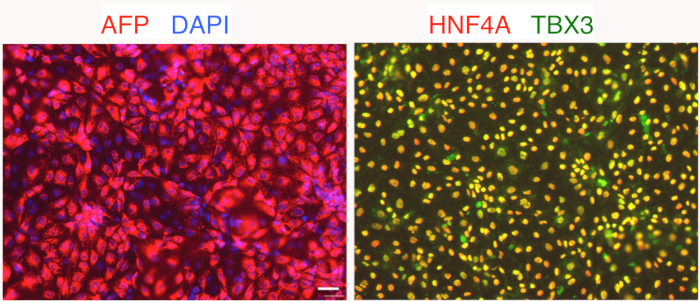
Figure 3: Immunostaining analysis of day 6 liver progenitors. hPSC were first differentiated into liver bud progenitors, which were immunostained for liver bud transcription factors HNF4A (red), TBX3 (green) as well as cytoplasmic liver bud marker AFP (red). Nuclei were counterstained with DAPI (blue) to assess total cell number. Scale bar = 50 μm. Please click here to view a larger version of this figure.
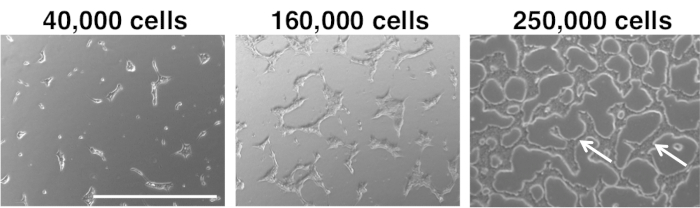
Figure 4: Cell seeding density of hPSCs. Low (left) and appropriate (middle and right) density of cells seeded. Scale bar = 1,000 μm. Arrow points to "bridges" between colonies of cells. Please click here to view a larger version of this figure.
| Base medium | Composition of base medium |
| CDM2 | 1:1 IMDM/F12, 0.1% m/v PVA, 1% concentrated lipids, 0.7 μg/mL human recombinant Insulin, 15 μg/mL Transferrin, 18 nM 1-thioglycerol |
| CDM3 | 1:1 IMDM/F12, 0.1% m/v PVA, 1% concentrated lipids, 10% KOSR |
| CDM4 | 1:1 IMDM/F12, 1% concentrated lipids, 15 μg/mL Transferrin |
| CDM5 | CMRL, 10% KOSR, 1% Glutamax |
Table 1: Composition of chemically defined media CDM2, CDM3, CDM4, and CDM5.
| Differentiation Stages | Duration | Factors | Doses | Base medium |
| Day 1 Primitive Streak | 1 day | Activin | 100 ng/mL | |
| CHIR99201 | 3 μM | CDM2 | ||
| PI103 | 50 nM | |||
| FGF2 | 10 ng/mL | |||
| Day 2 Definitive Endoderm | 1 day | Activin | 100 ng/mL | |
| DM3189 | 250 nM | CDM2 | ||
| PI103 | 50 nM | |||
| Day 3 Posterior Foregut | 1 day | A83-01 | 1 μM | |
| TTNPB | 75 nM | CDM3 | ||
| BMP4 | 30 ng/mL | |||
| FGF2 | 10 ng/mL | |||
| Day 6 Liver Bud progenitors | 2 days | Activin | 10 ng/mL | |
| C59 | 1 μM | CDM3 | ||
| BMP4 | 30 ng/mL | |||
| Forskolin | 1 μM | |||
| 1 day | Activin | 10 ng/mL | ||
| CHIR99201 | 1 μM | CDM3 | ||
| BMP4 | 30 ng/mL | |||
| Forskolin | 1 μM | |||
| Day 12 Hepatic progenitors | 6 days | BMP4 | 10 μg/mL | |
| OSM | 10 ng/mL | |||
| Dexamethasone | 10 μM | |||
| Forskolin | 10 μM | CDM4 | ||
| Ro4929097 | 2 μM | |||
| AA2P | 200 μg/mL | |||
| Insulin | 10 μg/mL | |||
| day 18 Hepatocytes | 6 days | Dexamethasone | 10 μM | |
| Forskolin | 10 μM | CDM4 or | ||
| Ro4929097 | 2 μM | CDM5 | ||
| AA2P | 200 μg/mL | |||
| Insulin | 10 μg/mL |
Table 2: Composition of differentiation media.
| Problem | Possible Reason | Proffered Solution |
| Cells in center of colony do not differentiate | i) Colony size was excessively large, cells in middle of large colonies were not accessible to differentiation signals | i) Check cell counting technqiue. |
| ii) Uneven distribution of clumps which merged together in the center of the well (or its periphery), forming very large colonies | ii) Shake the plate in a cross fashion to evenly distribute clumps during plating, and check under a microscope before placing it into the incubator | |
| iii) Cells received insufficient differentiation signals | iii) Add adequate volumes of differentiation media: add 1 mL of medium per well in a 12-well plate and 3 mL per well in a 6-well plate | |
| Poor efficiency of differentiation | i) Starting cell culture partially differentiated | i) Use a new, high-quality batch of undifferentiated hPSCs |
| ii) Colonies were too densely seeded, forming a confluent sheet of cells | ii) Seed cells and count cells accurately for differentiation, and shake evenly to distribute them | |
| iii) Residual media and unwanted signals were not washed off due to inadequate washing | iii) Residual mTeSR1 or induction media from the previous stage of differentiation will block differentiation; wash cells with IMDM prior to adding differentiation medium | |
| iv) Washing too harsh; inappropriate washing conditions will severely disrupt cell morphology | iv) Prior to adding either differentiation medium, wash briefly with IMDM. Use of DPBS (or media with different osmolarity or cold media) or extended washing, will compromise cell morphology and viability | |
| v) Prolonged or shortened period of differentiation stages, longer or shorter than recommended. | v) Adhere to the recommended timings for each stage of differentiation. |
Table 3: Potential problems and their possible causes and solutions.
Discussion
This method enables the generation of enriched populations of liver bud progenitors, and subsequently hepatocyte-like cells, from hPSCs. The ability to generate enriched populations of human liver cells is important for the practical utilization of such cells. Previous methods to generate hepatocytes from hPSCs yielded impure cell populations containing both liver and non-liver cells that, upon transplantation into rodents, yielded bone and cartilage in addition to liver tissue15. Hence the explicit suppression of non-liver differentiation is critical to generate enriched liver populations that might be suitable for a variety of applications.
Notably, controlled plating of hPSCs at the very first step is essential for efficient downstream differentiation. It is important to accurately plate hPSCs at a certain density for differentiation, and to evenly distribute these hPSCs across the well or plate during the process of plating. For example, for each well of a 12-well plate, seed 160,000 to 250,000 hPSCs per well; overall, it is imperative to titrate the cell seeding density and to ultimately test the cell density that is appropriate for each cell line (Figure 4). If too many hPSCs are seeded per well, they will form large colonies, which will adversely affect differentiation efficiencies, as cells in the middle of large colonies are less accessible to differentiation signals compared to cells near the periphery; colonies of heterogeneous sizes also will present similar issues. If cell densities are too low (e.g., below 200,000 cells/well of a 12-well plate), then there may not be sufficient material for differentiation and extensive cell death may be observed. The passaging and seeding method described above consistently generates hPSC clumps of suitable size for downstream differentiation.
One limitation of this method is that the hepatocyte-like cells generated are not identical to adult hepatocytes, as the hPSC-derived cells still express immature liver marker AFP. Moreover, CYP3A4 enzymatic activity is approximately 55 times lower in these hPSC-derived hepatocyte-like cells compared to primary adult human hepatocytes. A coming challenge will be to mature these hPSC-derived hepatocyte-like cells into fully-fledged, adult-like cells. A second limitation is that efficient differentiation is extremely dependent on the starting density of cells and therefore, it is very important to seed at the recommended density and to evenly disperse them across the plate (Table 3).
Overall, this protocol produces liver bud progenitors and hepatocyte-like cells at 89.0±3.1% purity in at least 3 hPSC lines. Second, hepatocyte-like cells expressed liver enzymes, secreted human ALBUMIN and expressed higher levels of liver genes than cells generated using extant differentiation approaches. Finally, the resultant hepatocyte-like cells not only exhibit certain hepatocyte functions in vitro, but most importantly they can function to some extent in vivo, as they can engraft a mouse model of chronic liver injury and improve short-term survival2.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Bing Lim for discussions and the Stanford Institute for Stem Cell Biology & Regenerative Medicine for infrastructure support. This work was supported by the California Institute for Regenerative Medicine (DISC2-10679) and the Stanford-UC Berkeley Siebel Stem Cell Institute (to L.T.A. and K.M.L.) and the Stanford Beckman Center for Molecular and Genetic Medicine as well as the Anonymous, Baxter and DiGenova families (to K.M.L.).
Materials
| Geltrex | Thermofisher Scientific | A1569601 | |
| 1:1 DMEM/F12 | Gibco | 11320033 | |
| 0.2 μm pore membrane filter | Millipore | GTTP02500 | |
| mTeSR1 | Stem Cell Technologies | 5850 | |
| Thiazovivin | Tocris Bioscience | 3845 | |
| Accutase | Gibco or Millipore | Gibco A11105, Millipore SCR005 | |
| IMDM, GlutaMAX™ Supplement | Thermofisher Scientific | 31980030 | |
| Ham's F-12 Nutrient Mix, GlutaMAX™ Supplement | Thermofisher Scientific | 31765035 | |
| KOSR, Knockout serum replacement | Thermofisher Scientific | 10828028 | |
| Poly(vinyl alcohol) | Sigma-Aldrich | P8136 | |
| Transferrin | Sigma-Aldrich | 10652202001 | |
| Chemically Defined Lipid Concentrate | Thermofisher Scientific | 11905031 | |
| human Activin | R&D | 338-AC | |
| CHIR99201 | Tocris | 4423 | |
| PI103 | Tocris | 2930/1 | |
| human FGF2 | R&D | 233-FB | |
| DM3189 | Tocris | 6053/10 | |
| A83-01 | Tocris | 2939/10 | |
| Human BMP4 | R&D | 314-BP | |
| C59 | Tocris | 5148 | |
| TTNPB | Tocris | 0761/10 | |
| Forskolin | Tocris | 1099/10 | |
| Oncostatin M | R&D | 295-OM | |
| Dexamethasone | Tocris | 1126 | |
| Ro4929097 | Selleck Chem | S1575 | |
| AA2P | Cayman chemicals | 16457 | |
| human recombinant Insulin | Sigma-Aldrich | 11061-68-0 |
References
- Bernal, W., Wendon, J. Acute Liver Failure. New England Journal of Medicine. 369, 2525-2534 (2013).
- Ang, L. T., et al. A Roadmap for Human Liver Differentiation from Pluripotent Stem Cells. Cell Reports. 22, 2190-2205 (2018).
- Fisher, R. A., Strom, S. C. Human Hepatocyte Transplantation: Worldwide Results. Transplantation. 82, 441-449 (2006).
- Dhawan, A. Clinical human hepatocyte transplantation: Current status and challenges. Liver transplantation. 21, S39-S44 (2015).
- Salama, H., et al. Autologous Hematopoietic Stem Cell Transplantation in 48 Patients with End-Stage Chronic Liver Diseases. Cell Transplantation. 19, 1475-1486 (2017).
- Tan, A. K. Y., Loh, K. M., Ang, L. T. Evaluating the regenerative potential and functionality of human liver cells in mice. Differentiation. 98, 25-34 (2017).
- Loh, K. M., et al. Efficient Endoderm Induction from Human Pluripotent Stem Cells by Logically Directing Signals Controlling Lineage Bifurcations. Cell Stem Cell. 14, 237-252 (2014).
- Loh, K. M., et al. Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell. , 451-467 (2016).
- Zhao, D., et al. Promotion of the efficient metabolic maturation of human pluripotent stem cell-derived hepatocytes by correcting specification defects. Cell Research. 23, 157-161 (2012).
- Si Tayeb, K., et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 51, 297-305 (2010).
- Carpentier, A., et al. Hepatic differentiation of human pluripotent stem cells in miniaturized format suitable for high-throughput screen. Stem Cell Research. 16, 640-650 (2016).
- Avior, Y., et al. Microbial-derived lithocholic acid and vitamin K2 drive the metabolic maturation of pluripotent stem cells-derived and fetal hepatocytes. Hepatology. 62, 265-278 (2015).
- Ogawa, S., et al. Three-dimensional culture and cAMP signaling promote the maturation of human pluripotent stem cell-derived hepatocytes. Development. 140, 3285-3296 (2013).
- Rostovskaya, M., Bredenkamp, N., Smith, A. Towards consistent generation of pancreatic lineage progenitors from human pluripotent stem cells. Philosophical Transactions of the Royal Society B: Biological Sciences. 370, 20140365 (2015).
- Haridass, D., et al. Repopulation Efficiencies of Adult Hepatocytes, Fetal Liver Progenitor Cells, and Embryonic Stem Cell-Derived Hepatic Cells in Albumin-Promoter-Enhancer Urokinase-Type Plasminogen Activator Mice. The American Journal of Pathology. 175, 1483-1492 (2009).

