Determination of Self- and Inter-(in)compatibility Relationships in Apricot Combining Hand-Pollination, Microscopy and Genetic Analyses
Summary
We present a methodology to establish the pollination requirements of apricot (Prunus armeniaca L.) cultivars combining the determination of self-(in)compatibility by fluorescence microscopy with the identification of the S-genotype by PCR analysis.
Abstract
Self-incompatibility in Rosaceae is determined by a Gametophytic Self-Incompatibility System (GSI) that is mainly controlled by the multiallelic locus S. In apricot, the determination of self- and inter-(in)compatibility relationships is increasingly important, since the release of an important number of new cultivars has resulted in the increase of cultivars with unknown pollination requirements. Here, we describe a methodology that combines the determination of self-(in)compatibility by hand-pollinations and microscopy with the identification of the S-genotype by PCR analysis. For self-(in)compatibility determination, flowers at balloon stage from each cultivar were collected in the field, hand-pollinated in the laboratory, fixed, and stained with aniline blue for the observation of pollen tube behavior under the fluorescence microscopy. For the establishment of incompatibility relationships between cultivars, DNA from each cultivar was extracted from young leaves and S-alleles were identified by PCR. This approach allows establishing incompatibility groups and elucidate incompatibility relationships between cultivars, which provides a valuable information to choose suitable pollinizers in the design of new orchards and to select appropriate parents in breeding programs.
Introduction
Self-incompatibility is a strategy of flowering plants to prevent self-pollination and promote outcrossing1. In Rosaceae, this mechanism is determined by a Gametophytic Self-Incompatibility System (GSI) that is mainly controlled by the multiallelic locus S2. In the style, the RNase gene encodes the S-stylar determinant, a RNase3, while a F-box protein, which determines the S-pollen determinant, is codified by the SFB gene4. The self-incompatibility interaction takes place through the inhibition of pollen tube growth along the style preventing the fertilization of the ovule5,6.
In apricot, a varietal renewal has taken place worldwide in the last two decades7,8. This introduction of an important number of new cultivars, from different public and private breeding programs, has resulted in the increase of apricot cultivars with unknown pollination requirements8.
Different methodologies have been used to determine pollination requirements in apricot. In the field, self-(in)compatibility may be established by controlled pollinations in caged trees or in emasculated flowers and subsequently recording the percentage of fruit set9,10,11,12. In addition, controlled pollinations have been carried out in the laboratory by semi-in vivo culture of flowers and analysis of the pollen tube behavior under fluorescence microscopy8,13,14,15,16,17. Recently, molecular techniques, such as PCR analysis and sequencing, have allowed the characterization of incompatibility relationships based on the study of the RNase and SFB genes18,19. In apricot, thirty-three S-alleles have been reported (S1 세스 S20, S22 세스 S30, S52, S53, Sv, Sx), including one allele related with self-compatibility (Sc)12,18,20,21,22,23,24. Up to now, 26 incompatibility groups have been stablished in this species according to the S-genotype8,9,17,25,26,27. Cultivars with the same S-alleles are inter-incompatible, whereas cultivars with at least one different S-allele and, consequently, allocated in different incompatible groups, are inter-compatible.
To define the pollination requirements of apricot cultivars, we describe a methodology that combines the determination of self-(in)compatibility by fluorescence microscopy with the identification of the S-genotype by PCR analysis in apricot cultivars. This approach allows establishing incompatibility groups and elucidate incompatibility relationships between cultivars.
Protocol
1. Self-(in)compatibility determination
- Sample the flowers in the field. It is necessary to collect the flowers at balloon stage (Figure 1A), corresponding to stage 58 on the BBCH scale for apricot28, to avoid unwanted previous pollination.
- Self- and cross-pollinations in the laboratory
- Remove the anthers of the flowers at balloon stage and place them on a piece of paper to dry at laboratory temperature.
- After 24 h, sieve the pollen grains by using a fine mesh (0.26 mm) (Figure 1B).
- Emasculate a group of 30 flowers at the same balloon stage for each self-pollination and cross-pollination and place the pistils on florist foam in water at laboratory temperature (Figure 1C).
- Hand pollinate the pistils with the help of a paintbrush with pollen from flowers of the same cultivar 24 h after emasculation. In addition, pollinate another set of pistils of each cultivar with pollen from flowers of a compatible pollinizer as control (Figure 1D).
- After 72 h, fix the pistils in a fixative solution of ethanol/acetic acid (3:1) for at least 24 h at 4 °C29. Then discard the fixative and add 75% ethanol ensuring that the samples are completely submerged in the solution. Samples can be conserved in this solution at 4 °C until use8,17,30,31,32.
- Evaluating pollen viability through in vitro pollen germination
- To prepare the germination medium, weight 25 g of sucrose, 0.075 g of boric acid (H3BO3) and 0.075 g of calcium nitrate (Ca(NO3)2)33.
- Add the components of the medium in 250 mL of distilled water and dissolve completely.
- Solidify the medium adding 2 g of agarose and mix by swirling.
- Check the pH of the medium using a pH meter and adjust the value to 7.0 with NaOH or HCl solution.
- Autoclave the mixture to sterilize the medium.
- After autoclaving, cool down the medium and distribute it into Petri dishes in a sterile laminar flow hood.
- Scatter the pollen grains of the same cultivars used for the controlled pollinations in the solidified pollen germination medium and observe them under the microscope after 24 h6.
NOTE: To sterilize the laminar flow hood, clean the surface with 70% ethanol and switch on the UV lamp during 10 min. - Store the Petri dishes in a refrigerator at 4 °C until use.
- Microscopy observations
- Wash the pistils three times for 1 h with distilled water and leave them in 5% sodium sulphite at 4 °C. After 24 h, autoclave them at 1 kg/cm2 during 10 min in sodium sulphite to soften the tissues34.
- Place the autoclaved pistils over a glass slide and, with the help of a scalpel, remove the trichomes around the ovary to get a better visualization of the pollen tubes. Then, squash the pistils with a cover glass.
- Prepare 0.1% (v/v) aniline blue stain: mix 0.1 mL of aniline blue in 100 mL of 0.1 N potassium phosphate tribasic (K3PO4). Apply a drop of aniline blue over the preparations to stain callose depositions during pollen tube growth.
- Observe the pollen tubes along the style by a microscope with UV epifluorescence using 340-380 bandpass and 425 longpass filters.
2. DNA extraction
- Sample 2-3 leaves in the field in spring. It is recommended to sample the leaves at young stages since DNA obtained is of higher quality and lower levels of phenolic compounds compared to old leaves.
- Extract Genomic DNA following the steps described in a commercially available kit (see Table of Materials).
- Analyze the quantity and quality of DNA concentrations using UV-vis spectrophotometer (260 nm).
3. S-allele identification
- Setting up of the PCR Reactions
- Prepare a 50 ng/μL dilution in distilled water of each DNA extraction sample.
- Thaw out the PCR reagents slowly and keep them on ice. Leave the DNA polymerase in the freezer until needed.
- Prepare the amplification reactions using the different combinations of primers. Create the PCR reaction mix by combining the components in Table 1. Vortex the PCR reaction mix well and distribute the volume indicated for the different combinations of primers to each well of the PCR plate. Then, add 1 μL of the DNA dilution in each well.
- Place the PCR plate in the thermocycler and run the corresponding PCR program shown in Table 1.
- Analyze the amplified fragments. There are mainly two different ways to analyze the PCR amplified fragments: capillary electrophoresis (CE) with fluorescent-labelled primers or as visualize amplicons of agarose gel electrophoresis with not-labelled primers.
- Capillary Electrophoresis
- To prepare the loading buffer, mix 35 μL of deionized formamide with 0.45 μL of labeled sizing standard. Vortex the reagent to mix well, and then dispense 35.5 μL into the well of the reader plate.
- Add 1 μL of the PCR product into the well. In addition, add a drop of mineral oil to prevent water evaporation.
- Prepare the separation plate adding separation buffer.
- Use the commercial software included with the gene analyzer (see Table of Materials). Create a new sample plate and save the sample names for all wells on the plate.
- Select the method of analysis. In this case, denature the samples at 90 °C for 120 s, inject at 2.0 kV for 30 s, and separate at 6.0 kV for 35 min.
- Insert the two plates into the gene analyzer. Fill the capillary array with distilled water.
- Load the patented linear polyacrylamide (LPA) gel. Finally, click Run.
- Gel Electrophoresis
- Prepare a 1% agarose gel adding 1.5 g of molecular biology grade agarose in 150 mL of 1x TAE (Tris-acetate-EDTA) electrophoresis running buffer (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA at pH 8.0). Dissolve the agarose by microwave heating for 2-3 min.
- To visualize the DNA, add 4 μL of a nucleic acid stain (see Table of Materials) and mix gently.
- Add a gel comb, with sufficient wells for ladders, controls and samples, into a gel tray. Then, pour slowly the mix into the middle of the gel tray and avoid bubbles.
- Let the gel cool down for 30-45 min at room temperature until the gel has completely solidified. Introduce the gel in the electrophoresis chamber, remove the gel comb and fill the chamber with enough 1x TAE buffer to cover the gel.
NOTE: Check the placement of the gel. The wells should be placed close to the negative pole since negatively charged DNA migrates towards the cathode. - Add 5 μL of loading buffer (0.1% (v/v) bromophenol blue) to the PCR products and mix well.
- To estimate the size of the bands, load 5 μL of DNA molecular weight ladder (see Table of Materials).
- Load the samples into the additional wells of the gel.
- Once all the samples and the DNA molecular weight ladder are loaded, run the gel at 90 V for 1-1.5 h, until the blue dye line is approximately at 75% the length of the gel.
- Visualize the bands in a transilluminator for nucleic acids.
- Capillary Electrophoresis
Representative Results
Pollination studies in apricot require the use of flowers at the late balloon stage one day before anthesis (Figure 1A). This stage is considered the most favorable for both pollen and pistil collection, since floral structures are nearly mature, but anther dehiscence has not yet occurred. This prevents the interference of undesired pollen, not only of pollen from the same flower but also from other flowers, since the closed petals impede the arrival of insects carrying external pollen. The pollen grains are easily sieved through a fine mesh (Figure 1B) from dehisced anthers previously placed on a piece of paper for 24 h at room temperature or with slight extra heat. Likewise, pistils are obtained from flowers at balloon stage after the removing of petals, sepals and stamens with the help of tweezers or fingernails (Figure 1C). Pistils can be self- and cross-pollinated with a fine brush (Figure 1D).
The hermaphroditic flowers of apricot have five dark red sepals, five white petals (Figure 1A), a single pistil (Figure 2A) and 25-30 stamens. The pistil has three main structures: stigma, style and ovary. The ovary has two ovules, and the fertilization of at least one of them is required for fruit setting. During pollination, insects, mainly bees, transfer pollen grains to the stigma (Figure 1A), where they germinate (Figure 2B) within 24 h following pollination. A pollen tube is produced from each germinating pollen grain, which grows through the pistil structures to reach the ovary after 3-4 days and fertilize one of the two ovules after around 7 days. In self-incompatible cultivars in which the S allele of the pollen grain is the same as one of the two of the pistils, pollen tube stops growing at the upper style, preventing fertilization (Figure 2C). However, the pollen tubes from a compatible cultivar, with a different S allele, can grow through the style (Figure 2D), reach the ovary (Figure 2E) and fertilize one of the two ovules.
The analysis of in vitro pollen germination showed good pollen viability in all the cultivars analyzed here, since most pollen tubes were longer than the length of the pollen grain after 24 h in the culture medium. Germinated pollen grains were observed at the stigma surface (Figure 2B) in pistils from all pollinations, indicating adequate pollination (Figure 3).
To determine the self-(in)compatibility for each cultivar, pollen tube behavior in self- and cross-pollinations done in laboratory-controlled conditions was observed under fluorescence microscopy. Pollen tube growth was recorded along the style in all the pistils examined. Cultivars were considered as self-incompatible when pollen tube growth was arrested along the style in most self-pollinated pistils (Figure 2C, Figure 3) and self-compatible when at least one pollen tube reached the base of the style in most of the pistils examined (Figure 2E, Figure 3).
The study of the S-locus by PCR analysis allowed characterizing the S-genotype of each cultivar. Firstly, the S-alleles were identified by the amplification of the first S-RNase intron using the primers SRc-F/SRc-R (Table 2). The size of the amplified fragments was analyzed by capillary electrophoresis (Figure 4A) and was used to classify the genotypes analyzed in their corresponding incompatibility group (I.G.) (Table 3).
Some pairs of alleles, such as S1 and S7 or S6 and S9, showed similar fragment sizes for the first intron. Thus, the differentiation of these alleles was done by amplifying a region of the second intron of the RNase with the primers Pru-C2/PruC4R, SHLM1/SHLM2 and SHLM3/SHLM4 (Table 2). The PruC2/PruC4R primer combination was used to distinguish between S6 and S9. For S6, a fragment of 1300 bp was amplified whereas a fragment of around 700 bp was observed for the S9 allele (Figure 4B, Table 3). The specific primers SHLM1/SHLM2 and SHLM3/SHLM4 amplified a fragment of approximately 650 bp in the S1 allele and 413 bp in the S7 allele (Figure 4C, Table 3).
The primers AprFBC8-(F/R) that amplify the V2 and HVb variable regions of the SFB gene were used to distinguish Sc and S8 alleles since both alleles show identical RNase sequence. The S8 allele showed a PCR-fragment of approximately 150 bp whereas a 500 bp fragment corresponded to the Sc allele (Figure 4D, Table 3). Once the S-genotype was determined for all the cultivars, self-incompatible cultivars were assigned to their corresponding incompatibility groups based on their S-alleles (Table 3).
This approach requires determining the self-(in)compatibility of each cultivar by controlled self- and cross-pollinations in the laboratory (Figure 5A) concomitantly with the characterization of the S-genotype by genetic analysis (Figure 5B). As a result, the pollination requirements of each cultivar and the incompatibility relationships among apricot cultivars can be determined.
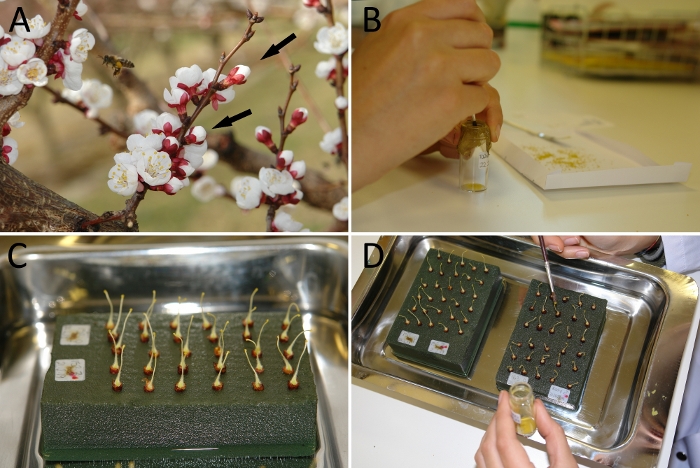
Figure 1. Experimental set up for the determination of self-(in)compatibility in apricot.
(A) Flowers at balloon stage (black arrows) in the field. (B) Sieve of pollen grains using a fine mesh. (C) Pistils placed on florist foam in water. (D) Hand-pollination of the pistils with the help of a paintbrush. Please click here to view a larger version of this figure.
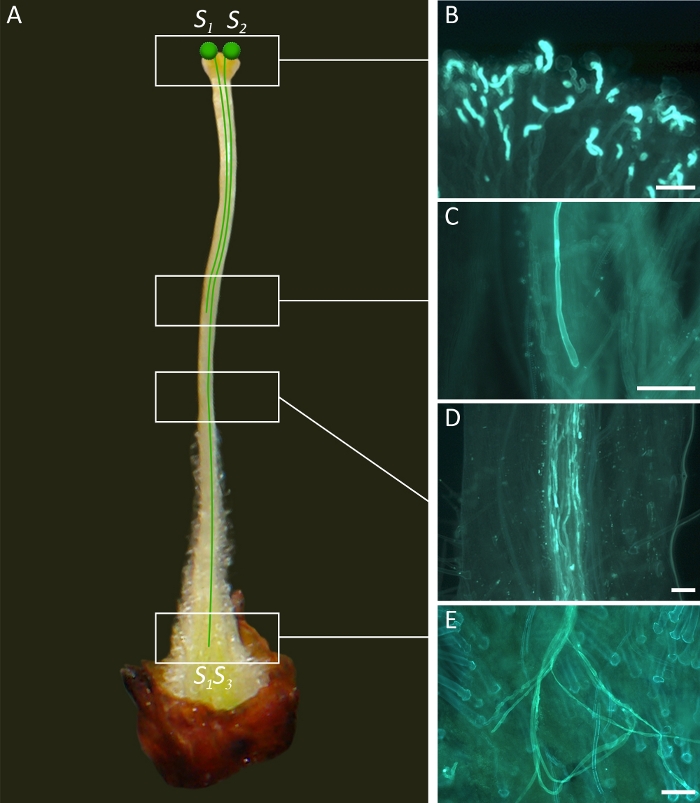
Figure 2. Diagrammatic representation of gametophytic incompatibility relationships in apricot flowers.
(A) In Gametophytic Self-Incompatibility (GSI), both compatible and incompatible pollen grains germinate on the stigma. The pollen grain carries one of two S-alleles of the original genotype, in this case either S1 or S2. If the S-allele of the pollen grain matches one of the two S-alleles of the pistil, in this case S1S3, pollen tube growth is inhibited in the upper one-third of the style. (B) Germination of pollen grains on the stigma surface. (C) Pollen tube arrested in the style indicating an incompatible behavior. (D) Pollen tubes growing along the style. (E) Pollen tubes at the base of the style indicating a compatible behavior. Scale bars, 100 μm. Please click here to view a larger version of this figure.
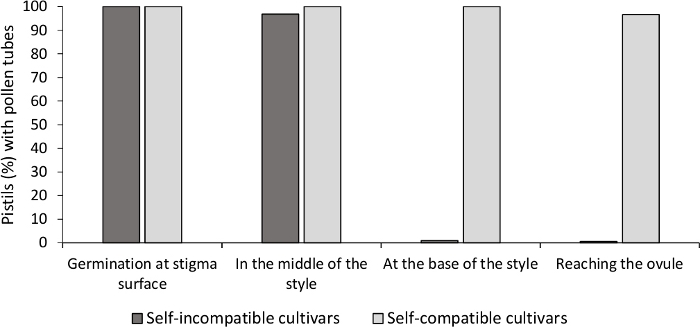
Figure 3. Representative results of pollen germination and pollen tube growth through the style for self-compatible and self-incompatible cultivars after self-pollinations.
Percentage of pistils with pollen grains germinating at the stigma surface, with pollen tubes in halfway the style, at the base of the style, and reaching the ovule. Please click here to view a larger version of this figure.
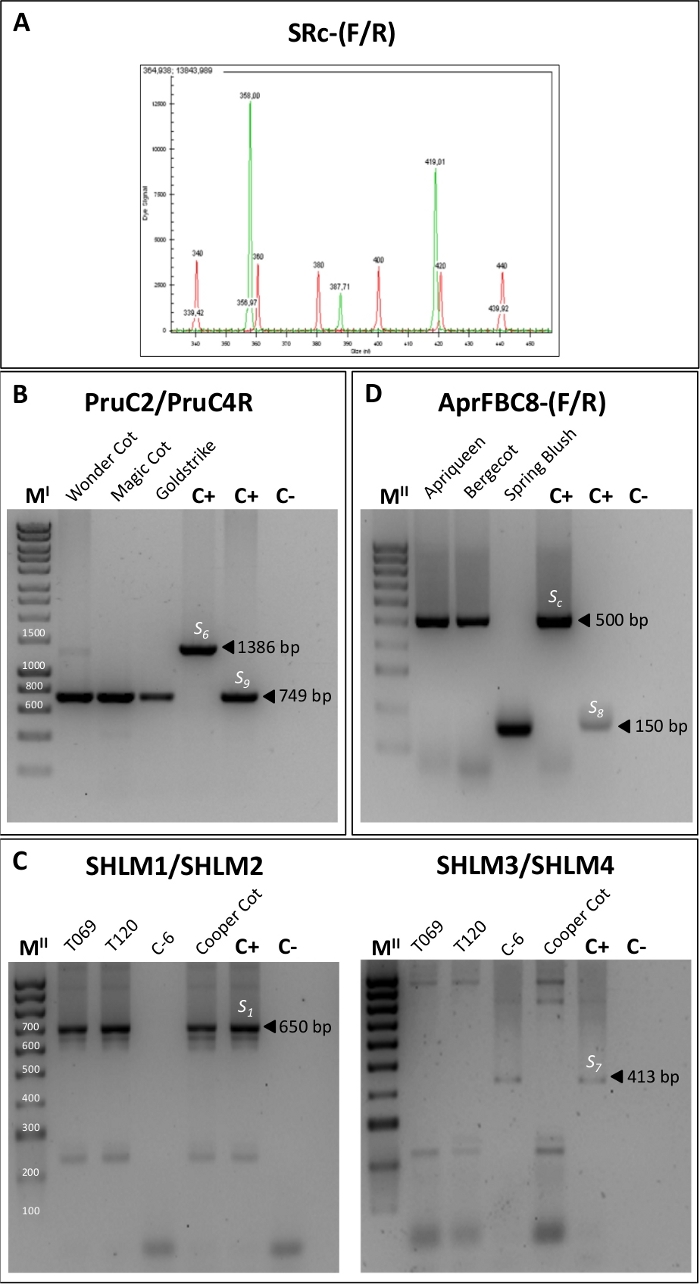
Figure 4. PCR fragment amplification using five primer pair combinations for the identification of S-alleles.
(A) Gene analyzer output for the SRc-(F/R) primers showing the size of the two amplified fragments of the RNase first intron region corresponding to the S-alleles. (B) PCR amplification using the primers PruC2/PruC4R for the identification of the S6 and S9 alleles. (C) PCR products obtained using the specific primers SHLM1 and SHLM2 for the differentiation of the S1 allele and SHLM3 and SHLM4 to distinguish the S7 allele. (D) PCR amplification with the AprFBC8-(F/R) primers for identifying Sc and S8 alleles. MI: 1 kb DNA Ladder. MII: 100 bp DNA Ladder. Please click here to view a larger version of this figure.
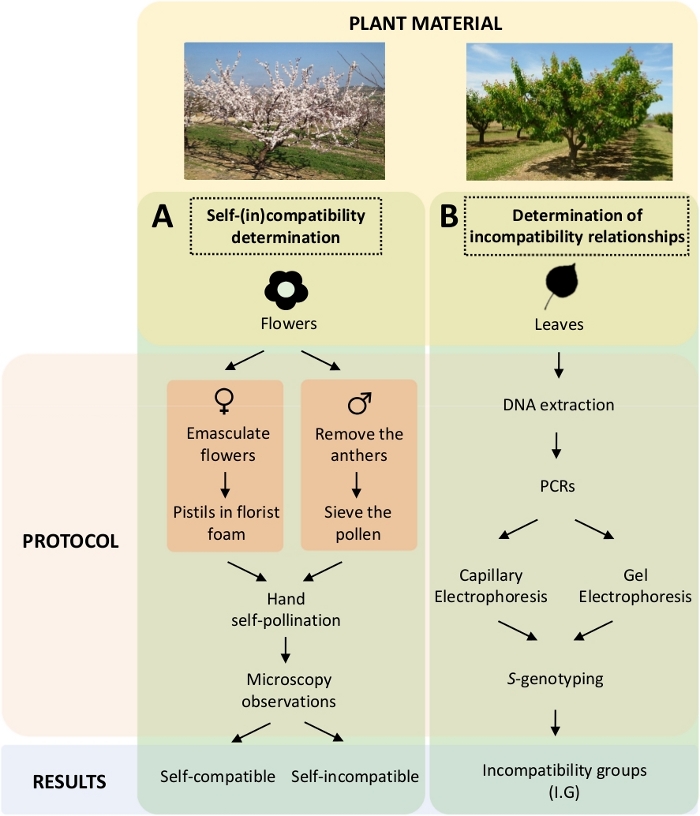
Figure 5. Scheme of the experimental design to elucidate the self- and inter-(in) compatibility relationships in apricot cultivars.
(A) Workflow of self-(in)compatibility determination by controlled pollinations in the laboratory. (B) Workflow of the S-allele identification by molecular approaches. Please click here to view a larger version of this figure.
| PCR Master Mix | Thermocycler conditions | ||||||
| Components | Final Concentration | 15 μL reaction | Cycle Step | Temperature | Time | Cycles | |
| 10x NH4 Reaction Buffer | 10x | 1.5 μL | Initial denaturation | 94 °C | 3 min | 1 | |
| 50 mM MgCl2 Solution | 25 mM | 1.2 μL | Denaturing | 94 °C | 1 min | 35 | |
| 100 mM dNTP | 2.5 mM | 0.6 μL | Annealing | 55 °C | 1 min | ||
| Primer SRc-F | 10 μM | 0.6 μL | Extension | 72 °C | 3 min | ||
| Primer SRc-R | 10 μM | 0.6 μL | Final Extension | 72 °C | 5 min | 1 | |
| 500 U Taq DNA Polymerase | 0.5 U | 0.2 μL | 4 °C | hold | |||
| H2O | 8.3 μL | ||||||
| Components | Final Concentration | 25 μL reaction | Cycle Step | Temperature | Time | Cycles | |
| 10x PCR buffer | 10x | 2.5 μL | Initial denaturation | 94 °C | 2 min | 1 | |
| 5x Q-solution | 5x | 5 μL | Denaturing | 94 °C | 10 s | 10 | |
| 100 mM dNTP | 2.5 mM | 0.5 μL | Annealing | 55 °C | 2 min | ||
| Primer PruC2 | 10 μM | 0.2 μL | Extension | 68 °C | 2 min | ||
| Primer C4R | 10 μM | 0.2 μL | Denaturing | 94 °C | 10 s | 25 | |
| 250 U Taq DNA Polymerase | 10 U | 0.13 μL | Annealing | 58 °C | 2 min | ||
| H2O | 15.5 μL | Extension* | 68 °C | 2 min | |||
| Final Extension | 72 °C | 5 min | 1 | ||||
| 4 °C | hold | ||||||
| * with 10 s added each cycle to the 68 %C extension step. | |||||||
| Components | Final Concentration | 25 μL reaction | Cycle Step | Temperature | Time | Cycles | |
| 10x PCR buffer | 10x | 2.5 μL | Initial denaturation | 94 °C | 2 min | 1 | |
| 5x Q-solution | 5x | 5 μL | Denaturing | 94 °C | 30 s | 35 | |
| 100 mM dNTP | 2.5 mM | 0.5 μL | Annealing | 62 °C | 1.5 min | ||
| Primer SHLM1 | 10 μM | 0.2 μL | Extension | 72 °C | 2 min | ||
| Primer SHLM2 | 10 μM | 0.2 μL | Final Extension | 72 °C | 5 min | 1 | |
| 250 U Taq DNA Polymerase | 10 U | 0.13 μL | 4 °C | hold | |||
| H2O | 15.5 μL | ||||||
| Components | Final Concentration | 20 μL reaction | Cycle Step | Temperature | Time | Cycles | |
| 5x PCR Buffer | 5x | 4 μL | Initial denaturation | 98 °C | 30 s | 1 | |
| dNTP | 2.5 mM | 1.6 μL | Denaturing | 98 °C | 10 s | 35 | |
| Primer SHLM3 | 10 μM | 1 μL | Annealing | 51 °C | 30 s | ||
| Primer SHLM4 | 10 μM | 1 μL | Extension | 72 °C | 1 min | ||
| 100 U DNA Polymerase | 5 U | 0.2 μL | Final Extension | 72 °C | 5 min | 1 | |
| H2O | 12.4 μL | 4 °C | hold | ||||
| Components | Final Concentration | 25 μL reaction | Cycle Step | Temperature | Time | Cycles | |
| 10x PCR buffer | 10x | 2.5 μL | Initial denaturation | 94 °C | 2 min | 1 | |
| 100 mM dNTP | 2.5 mM | 2 μL | Denaturing | 94 °C | 30 s | 35 | |
| Primer FBC8-F | 10 μM | 1 μL | Annealing | 55 °C | 1.5 min | ||
| Primer FBC8-R | 10 μM | 1 μL | Extension | 72 °C | 2 min | ||
| 250 U Taq DNA Polymerase | 10 U | 0.125 μL | Final Extension | 72 °C | 5 min | 1 | |
| H2O | 17.4 μL | 4 °C | hold | ||||
Table 1. Reaction and cycling conditions for different primer combinations used in this protocol.
| Primers | Sequence | Reference |
| SRc-F | 5'-CTCGCTTTCCTTGTTCTTGC-3' | 18 |
| SRc-R | 5'-GGCCATTGTTGCACCCCTTG-3' | 18 |
| Pru-C2 | 5'-CTTTGGCCAAGTAATTATTCAAACC-3' | 35 |
| Pru-C4R | 5'-GGATGTGGTACGATTGAAGCG-3' | 35 |
| SHLM1-F | 5'-GGTGGAGGTGATAAGGTAGCC-3' | 17 |
| SHLM2-R | 5'-GGCTGCATAAGGAAGCTGTAGG-3' | 17 |
| SHLM3-F | 5'-TATATCTTACTCTTTGGC-3' | 17 |
| SHLM4-R | 5'-CACTATGATAATGTGTATG-3' | 17 |
| AprFBC8-F | 5'-CATGGAAAAAGCTGACTTATGG-3' | 26 |
| AprFBC8-R | 5'-GCCTCTAATGTCATCTACTCTTAG-3' | 26 |
Table 2. Primers used in this protocol, sequence and reference for S-genotype characterization in Prunus armeniaca.
| Cultivar | SRc-(F/R) (bp) | PruC2/PruC4R (bp) | SHLM1/SHLM2 (bp) | SHLM3/SHLM4 (bp) | AprFBC8-(F/R) (bp) | S-Genotype | Incompatibility group (I.G) |
| Wonder Cot8 | 420, 420 | 749, 1386 | S6S9 | VIII | |||
| Magic Cot8 | 334, 420 | 749 | S2S9 | XX | |||
| Goldstrike8 | 334, 420 | 749 | S2S9 | – | |||
| T06917 | 334, 408 | 650 | S1S2 | I | |||
| T12017 | 334, 408 | 650 | S1S2 | – | |||
| C-6 | 334, 408 | 413 | S2S7 | IV | |||
| Cooper Cot8 | 274, 408 | 650 | S1S3 | XVIII | |||
| Apriqueen | 358, 358 | 500 | ScSc | – | |||
| Bergecot8 | 334, 358 | 500 | S2Sc | – | |||
| Spring Blush8 | 274, 358 | 150 | S3S8 | XXI |
Table 3. S-genotyping of apricot cultivars with five primer pairs used in this protocol and incompatibility group assignment. The different polymerase chain reaction product sizes of S-alleles amplified using SRc-(F/R), PruC2/PruC4R, SHLM1/ SHLM2, SHLM3/SHLM4, and AprFBC8-(F/R) primers are shown in the table.
Discussion
Traditionally, most commercial apricot European cultivars were self-compatible36. Nevertheless, the use of North American self-incompatible cultivars as parents in breeding programs in the last decades has resulted in the release of an increasing number of new self-incompatible cultivars with unknown pollination requirements7,8,37. Thus, the determination of self- and inter-(in)compatibility relationships in apricot cultivars is increasingly important. This is accentuated in those areas where winter chilling is decreasing, since high year to year variations in the time of flowering are preventing the coincidence in flowering of cultivars and their pollenizers in many cases, especially in cultivars with high chilling requirements38. The methodology described herein, combining hand-pollination, microscopy and genetic analyses has been very useful to determine the self(in)compatibility of each cultivar and to establish its potential pollinizer cultivars.
Pollination requirements can be determined through field-control experiments in orchard conditions11,39. However, the exposition to external factors including meteorological adverse conditions can cause pollination failure10, which may result in erroneous diagnoses of self-incompatibility. The methodology described herein allows to evaluate self-(in)compatibility more accurately by microscopy observations of hand-pollinated flowers in laboratory-controlled conditions, avoiding environmental influence. Moreover, this approach allows analyzing a higher number of cultivars per year, since only a small number of flowers is required instead of several adult trees for each cultivar that are required in field experiments40.
Incompatibility relationships can be established combining hand-pollinations and microscopy14. However, pollinations can only be performed for a short period during the flowering season in spring, and adult trees near the laboratory are needed, since the lifespan of the flowers collected is very short. Thus, the number of incompatibility relationships that can be analyzed by controlled hand-pollinations in each season is very low. The characterization of the genes encoded by the S-locus has enabled the development of PCR-based methods for S-allele genotyping18,41. This approach accelerates S-allele identification since it does not require flowers, and the experiments can be carried out with any vegetative tissue42. This extends the period during which plant material, usually young leaves, can be collected43. Furthermore, the leaves can be lyophilized or frozen, so that the analysis can be done at any time of the year, unlike pollinations that can only be done on fresh flowers during the flowering season44. An additional benefit is that leaves can be collected from young trees even before entering flowering age, facilitating the collection of samples and the early obtaining of results45.
The genetic analysis allows a better differentiation of self-incompatibility alleles since it provides precise results of amplified fragment sizes21,46. To date, thirty-three S-alleles have been identified in apricot12,18,20,21,22,23,24, which has allowed to establish 36 incompatibility groups based on S-genotype8,9,17,25,26,27. On the other hand, a drawback of this methodology is that different alleles in the same range size or mutations can be erroneously identified as the same allele. Thus, Sc and S8 alleles are identical for the RNase sequence but a 358-bp insertion is found in the SFB gene of Sc19. Likewise, the first intron region of the alleles S1 and S7 are identical and are indistinguishable using the primers SRc-F/SRc-R. In addition, several homologies, such as S6 and S528 or S20 and S55, and S7, S13 (EF062341) and S4617, have been found because some of these alleles have been partially sequenced or by failures during PCR amplification and, consequently, further work is needed to distinguish them correctly.
PCR analysis and S-RNase sequencing are adequate for establishing incompatibility relationships through the identification of S-alleles and the allocation of cultivars in their corresponding Incompatibility Group8,17,26,27. However, this methodology has the limitation of preventing the determination of the self-(in)compatibility for particular apricot cultivars. Self-compatibility (SC) has been associated to particular S-alleles in other Prunus species47, as almond (Sf)48,49 or sweet cherry (S4’)50,51. However, in apricot, the Sc allele, which has been associated to SC21, can be erroneously identified as S8, a self-incompatible allele19,22, and possible mutations not linked to the S locus, as the M-locus12,52, conferring SC have been identified. Recently, the M-locus has been genotyped using SSR markers12. Therefore, the genetic identification of SC for apricot genotypes needs further research and, in order to avoid mistakes due to factors not linked to the S locus, in this work the characterization of self-(in)compatibility has been determined also by phenotyping the behavior of the pollen tubes through the pistil of self-pollinated flowers.
The methodology described herein combining the determination of self-(in)compatibility by hand-pollinations in laboratory conditions with the subsequent observation of the behavior of pollen tubes in the pistil of controlled self-pollinations under the fluorescence microscopy and the identification of the S-genotype by PCR analysis allows establishing the pollination requirements of apricot cultivars. This provides a valuable information for growers and breeders, since it allows establishing the incompatibility relationships between cultivars to choose suitable pollinizers in the design of new orchards as well as to select appropriate parents to design new crosses in apricot breeding programs.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was funded by Ministerio de Ciencia, Innovación y Universidades-European Regional Development Fund, European Union (AGL2016-77267-R, and AGL2015-74071-JIN); Instituto Nacional de Investigación y Tecnología Agraria y Alimentaria (RFP2015-00015-00, RTA2017-00003-00); Gobierno de Aragón-European Social Fund, European Union (Grupo Consolidado A12_17R), Fundación Biodiversidad, and Agroseguro S.A.
Materials
| Agarose D1 Low EEO | Conda | 8010.22 | |
| BIOTAQ DNA Polymerase kit | Bioline | BIO-21060 | |
| Bright field microscope | Leica Microsystems | DM2500 | |
| CEQ System Software | Beckman Coulter | ||
| DNeasy Plant Mini Kit | QIAGEN | 69106 | |
| dNTP Set, 4 x 25 µmol | Bioline | BIO-39025 | |
| GenomeLab DNA Size Standard Kit – 400 | Beckman Coulter | 608098 | |
| GenomeLab GeXP Genetic Analysis System | Beckman Coulter | ||
| GenomeLab Separation Buffer | Beckman Coulter | 608012 | |
| GenomeLab Separation Gel LPA-1 | Beckman Coulter | 391438 | |
| HyperLadder 100bp | Bioline | BIO-33029 | |
| HyperLadder 1kb | Bioline | BIO-33025 | |
| Image Analysis System | Leica Microsystems | ||
| Molecular Imager VersaDoc MP 4000 system | Bio-Rad | 170-8640 | |
| NanoDrop One Spectrophotometer | Thermo Fisher Scientific | 13-400-518 | |
| pH-Meter BASIC 20 | Crison | ||
| Phusion High-Fidelity PCR Kit | Thermo Fisher Scientific | F553S | |
| Power Pack P 25 T | Biometra | ||
| Primer Forward | Isogen Life Science | ||
| Primer Reverse | Isogen Life Science | ||
| Quantity One Software | Bio-Rad | ||
| Stereoscopic microscope | Leica Microsystems | MZ-16 | |
| Sub-Cell GT | Bio-Rad | ||
| SYBR Safe DNA Gel Stain | Thermo Fisher Scientific | S33102 | |
| T100 Thermal Cycler | Bio-Rad | 1861096 | |
| Taq DNA Polymerase | QIAGEN | 201203 | |
| Vertical Stand Autoclave | JP Selecta |
References
- Silva, N. F., Goring, D. R. Mechanisms of self-incompatibility in flowering plants. Cellular and Molecular Life Sciences. 58, 1988-2007 (2001).
- Charlesworth, D., Vekemans, X., Castric, V., Glémin, S. Plant self-incompatibility systems: A molecular evolutionary perspective. New phytologist. 168, 61-69 (2005).
- Tao, R., et al. Identification of stylar RNases associated with gametophytic self-incompatibility in almond (Prunus dulcis). Plant and Cell Physiology. 38, 304-311 (1997).
- Ushijima, K., et al. Structural and transcriptional analysis of the self-incompatibility locus of almond: Identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. The Plant cell. 15, 771-781 (2003).
- Bedinger, P. A., Broz, A. K., Tovar-Mendez, A., McClure, B. Pollen-Pistil Interactions and Their Role in Mate Selection. Plant Physiology. 173, 79-90 (2017).
- Guerra, M. E., Rodrigo, J. Japanese plum pollination: A review. Scientia Horticulturae. 197, 674-686 (2015).
- Zhebentyayeva, T., Ledbetter, C., Burgos, L., Llacer, G., Badenes, M. L., Byrne, D. Apricot. Fruit Breeding. , 415-458 (2012).
- Herrera, S., Lora, J., Hormaza, J. I., Herrero, M., Rodrigo, J. Optimizing Production in the New Generation of Apricot Cultivars: Self-incompatibility, S-RNase Allele Identification, and Incompatibility Group Assignment. Frontiers in Plant Science. 9, 527 (2018).
- Egea, J., Burgos, L. Detecting Cross-incompatibility of Three North American Apricot Cultivars and Establishing the First Incompatibility Group in Apricot. Journal of the American Society for Horticultural Science. 121, 1002-1005 (1996).
- Rodrigo, J., Herrero, M. Effects of pre-blossom temperatures on flower development and fruit set in apricot. Scientia Horticulturae. 92, 125-135 (2002).
- Julian, C., Herrero, M., Rodrigo, J. Flower bud differentiation and development in fruiting and non-fruiting shoots in relation to fruit set in apricot (Prunus armeniaca). Trees. 24, 833-841 (2010).
- Muñoz-Sanz, J. V., Zuriaga, E., López, I., Badenes, M. L., Romero, C. Self-(in)compatibility in apricot germplasm is controlled by two major loci, S and M. BMC Plant Biology. 17, 82 (2017).
- Burgos, L., Berenguer, T., Egea, J. Self- and Cross-compatibility among Apricot Cultivars. HortScience. 28, 148-150 (1993).
- Rodrigo, J., Herrero, M. Evaluation of pollination as the cause of erratic fruit set in apricot “Moniqui”. Journal of Horticultural Science. 71, 801-805 (1996).
- Milatović, D., Nikolić, D., Krška, B. Testing of self-(in)compatibility in apricot cultivars from European breeding programmes. Horticultural Science. 40 (2), 65-71 (2013).
- Milatović, D., Nikolić, D., Fotirić-Aksić, M., Radović, A. Testing of self-(in)compatibility in apricot cultivars using fluorescence microscopy. Acta Scientiarum Polonorum, Hortorum Cultus. 12 (6), 103-113 (2013).
- Herrera, S., Rodrigo, J., Hormaza, J. I., Lora, J. Identification of Self-Incompatibility Alleles by Specific PCR Analysis and S-RNase Sequencing in Apricot. Int J Mol Sci. 19, 3612 (2018).
- Romero, C., et al. Analysis of the S-locus structure in Prunus armeniaca L. Identification of S-haplotype specific S-RNase and F-box genes. Plant Molecular Biology. 56, 145-157 (2004).
- Halász, J., Pedryc, A., Hegedus, A. Origin and dissemination of the pollen-part mutated SC haplotype which confers self-compatibility in apricot (Prunus armeniaca). New Phytologist. 176, 792-803 (2007).
- Halász, J., Hegedus, A., Hermán, R., Stefanovits-Bányai, &. #. 2. 0. 1. ;., Pedryc, A. New self-incompatibility alleles in apricot (Prunus armeniaca L.) revealed by stylar ribonuclease assay and S-PCR analysis. Euphytica. 145, 57-66 (2005).
- Vilanova, S., Romero, C., Llacer, G., Badenes, M. L., Burgos, L. Identification of Self-(in)compatibility Alleles in Apricot by PCR and Sequence Analysis. Journal of the American Society for Horticultural Science. 130, 893-898 (2005).
- Feng, J., et al. Detection and transcript expression of S-RNase gene associated with self-incompatibility in apricot (Prunus armeniaca L.). Molecular Biology Reports. 33, 215-221 (2006).
- Zhang, L., et al. Identification of self-incompatibility (S-) genotypes of Chinese apricot cultivars. Euphytica. 160, 241-248 (2008).
- Wu, J., et al. Identification of S-haplotype-specific S-RNase and SFB alleles in native Chinese apricot (Prunus armeniaca L). Journal of Horticultural Science and Biotechnology. 84, 645-652 (2009).
- Szabó, Z., Nyéki, J. Blossoming, fructification and combination of apricot varieties. Acta Horticulturae. 293, 295-302 (1991).
- Halász, J., Pedryc, A., Ercisli, S., Yilmaz, K. U., Hegedűs, A. S-genotyping supports the genetic relationships between Turkish and Hungarian apricot germplasm. Journal of the American Society for Horticultural Science. 135, 410-417 (2010).
- Lachkar, A., et al. Identification of self-(in)compatibility S-alleles and new cross-incompatibility groups in Tunisian apricot (Prunus armeniaca L.) cultivars. The Journal of Horticultural Science and Biotechnology. 88, 497-501 (2013).
- Pérez-Pastor, A., Ruiz-Sánchez, M. C., Domingo, R., Torrecillas, A. Growth and phenological stages of Búlida apricot trees in South-East. Agronomie. 24, 93-100 (2004).
- Williams, J. H., Friedman, W. E., Arnold, M. L. Developmental selection within the angiosperm style: using gamete DNA to visualize interspecific pollen competition. Proceedings of the National Academy of Sciences of the United States of America. 96, 9201-9206 (1999).
- Julian, C., Herrero, M., Rodrigo, J. Anther meiosis time is related to winter cold temperatures in apricot (Prunus armeniaca L.). Environmental and Experimental Botany. 100, 20-25 (2014).
- Guerra, M. E., López-Corrales, M., Wünsch, A., Rodrigo, J. Lack of Fruit Set Caused by Ovule Degeneration in Japanese Plum. Journal of the American Society for Horticultural Science. 136 (6), 375-381 (2011).
- Guerra, M. E., Wünsch, A., López-Corrales, M., Rodrigo, J. Flower Emasculation as the Cause for Lack of Fruit Set in Japanese Plum Crosses. Journal of the American Society for Horticultural Science. 135 (6), 556-562 (2010).
- Hormaza, J. I., Pinney, K., Polito, V. S. Correlation in the tolerance to ozone between sporophytes and male gametophytes of several fruit and nut tree species (Rosaceae). Sexual Plant Reproduction. 9, 44-48 (1996).
- Alcaraz, M. L., Hormaza, J. I., Rodrigo, J. Pistil Starch Reserves at Anthesis Correlate with Final Flower Fate in Avocado (Persea americana). PLOS ONE. 8 (10), 78467 (2013).
- Tao, R., et al. Molecular typing of S-alleles through Identification, Characterization and cDNA cloning for S-RNases in Sweet Cherry. Journal of the American Society for Horticultural Science. 124, 224-233 (1999).
- Burgos, L., et al. The self-compatibility trait of the main apricot cultivars and new selections from breeding programmes. Journal of Horticultural Science. 72, 147-154 (1997).
- Hormaza, J. I., Yamane, H., Rodrigo, J., Kole, C. Apricot. Genome Mapping and Molecular Breeding in Plants, Volume 4 Fruits and Nuts. , 171-187 (2007).
- Benmoussa, H., Ghrab, M., Ben Mimoun, M., Luedeling, E. Chilling and heat requirements for local and foreign almond (Prunus dulcis Mill.) cultivars in a warm Mediterranean location based on 30 years of phenology records. Agricultural and Forest Meteorology. 239, 34-46 (2017).
- Rodrigo, J., Herrero, M., Hormaza, J. I. Pistil traits and flower fate in apricot (Prunus armeniaca). Annals of Applied Biology. 154, 365-375 (2009).
- Williams, R. R., Williams, R. R., Wilson, D. Techniques used in fruit-set experiments. Towards Regulated Cropping. , 57-61 (1970).
- Sutherland, B. G., Robbins, T. P., Tobutt, K. R. Primers amplifying a range of Prunus S-alleles. Plant Breeding. 123, 582-584 (2004).
- Murray, M. G., Thompson, W. F. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Research. 8, 4321-4325 (1980).
- Porebski, S., Bailey, L. G., Baum, B. R. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant Molecular Biology Reporter. 15 (1), 8-15 (1997).
- Rogers, S. O., Bendich, A. J. Extraction of DNA from milligram amounts of fresh, herbarium and mummified plant tissues. Plant Molecular Biology. 5 (2), 69-76 (1985).
- Hormaza, J. I. Molecular characterization and similarity relationships among apricot (Prunus armeniaca L.) genotypes using simple sequence repeats. Theoretical and Applied Genetics. 104, 321-328 (2002).
- Sonneveld, T., Tobutt, K. R., Robbins, T. P. Allele-specific PCR detection of sweet cherry self-incompatibility (S) alleles S1 to S16 using consensus and allele-specific primers. Theoretical and Applied Genetics. 107, 1059-1070 (2003).
- Hegedus, A., Lénárt, J., Halász, J. Sexual incompatibility in Rosaceae fruit tree species: molecular interactions and evolutionary dynamics. Biologia Plantarum. 56 (2), 201-209 (2012).
- Fernández i Martí, A., Gradziel, T. M., Socias i Company, R. Methylation of the Sf locus in almond is associated with S-RNase loss of function. Plant Molecular Biology. 86, 681-689 (2014).
- Company, R. S. i., Kodad, O., Martí, A. F. i., Alonso, J. M. Mutations conferring self-compatibility in Prunus species: From deletions and insertions to epigenetic alterations. Scientia Horticulturae. 192, 125-131 (2015).
- Boskovic, R., Tobutt, K. R. Correlation of stylar ribonuclease zymograms with incompatibility alleles in sweet cherry. Euphytica. 90, 245-250 (1996).
- Cachi, A. M., Wünsch, A. S-genotyping of sweet cherry varieties from Spain and S-locus diversity in Europe. Euphytica. 197 (2), 229-236 (2014).
- Zuriaga, E., et al. An S-locus Independent Pollen Factor Confers Self-Compatibility in “Katy” Apricot. PLoS ONE. 8 (1), 53947 (2013).

