Longitudinal Follow-Up of Urinary Tract Infections and Their Treatment in Mice using Bioluminescence Imaging
Summary
This manuscript describes the intravesical administration of uropathogenic bacteria with a lux operon to induce a urinary tract infection in mice and subsequent longitudinal in vivo analysis of the bacterial load using bioluminescence imaging.
Abstract
Urinary tract infections (UTI) rank among the most common bacterial infections in humans and are routinely treated with empirical antibiotics. However, due to increasing microbial resistance, the efficacy of the most used antibiotics has declined. To find alternative treatment options, there is a great need for a better understanding of the UTI pathogenesis and the mechanisms that determine UTI susceptibility. In order to investigate this in an animal model, a reproducible, non-invasive assay to study the course of UTI is indispensable.
For years, the gold standard for the enumeration of bacterial load has been the determination of Colony Forming Units (CFU) for a particular sample volume. This technique requires post-mortem organ homogenates and serial dilutions, limiting data output and reproducibility. As an alternative, bioluminescence imaging (BLI) is gaining popularity to determine the bacterial load. Labeling pathogens with a lux operon allow for the sensitive detection and quantification in a non-invasive manner, thereby enabling longitudinal follow-up. So far, the adoption of BLI in UTI research remains limited.
This manuscript describes the practical implementation of BLI in a mouse urinary tract infection model. Here, a step-by-step guide for culturing bacteria, intravesical instillation and imaging is provided. The in vivo correlation with CFU is examined and a proof-of-concept is provided by comparing the bacterial load of untreated infected animals with antibiotic-treated animals. Furthermore, the advantages, limitations, and considerations specific to the implementation of BLI in an in vivo UTI model are discussed. The implementation of BLI in the UTI research field will greatly facilitate research on the pathogenesis of UTI and the discovery of new ways to prevent and treat UTI.
Introduction
Urinary tract infections (UTI) are among the most common bacterial infections in humans. Almost half of all women will experience a symptomatic UTI during their lifetime1. Infections limited to the bladder can give rise to urinary symptoms such as increase in urinary frequency, urgency, hematuria, incontinence, and pain. When the infection ascends to the upper urinary tract, patients develop pyelonephritis, with malaise, fever, chills, and back pain. Furthermore, up to 20% of patients with UTI suffer from recurrent infections resulting in a dramatic decrease in antibiotic sensitivity2,3,4. In recent years, there has been a growing interest in novel therapies for the treatment and prevention of recurrent UTI. Despite a better understanding of the innate and adaptive immunity of the lower urinary tract and of the bacterial virulence factors necessary for invasion and colonization, no radical changes in the treatment regime have been translated to the daily urological practice2. In order to study UTI pathogenesis and susceptibility in vivo, a reproducible and non-invasive assay is indispensable.
Multiple animal UTI models have been described ranging from nematodes to primates, but the murine model is predominantly used5,6. This model consists of transurethral catheterization of (female) mice and subsequent instillation of a bacterial suspension, most commonly uropathogenic Escherichia coli (UPEC), directly into the bladder lumen7. After inoculation, the bacterial load has traditionally been quantified by determining colony forming units (CFU). This technique requires sacrificing animals to obtain post-mortem organ homogenates and serial dilutions, limiting data output and reproducibility. Moreover, longitudinal follow-up of the bacterial load in individual animals is not possible using this technique.
In 1995, Contag et al. suggested the use of bioluminescent-tagged pathogens to monitor disease processes in living animals8,9. Since then, bioluminescence imaging (BLI) has been applied to numerous infection models such as meningitis, endocarditis, osteomyelitis, skin, and soft tissue infections, etc.10,11,12. In the murine UTI model, a UPEC strain with the complete lux operon (luxCDABE) from Photorhabdus luminescens can be used13. An enzymatic reaction is catalyzed by the bacterial luciferase which is dependent on the oxidation of long-chain aldehydes reacting with reduced flavin mononucleotide in the presence of oxygen, yielding the oxidized flavin, a long-chain fatty acid and light12. The lux operon encodes for the luciferase and other enzymes required for the synthesis of the substrates. Therefore, all metabolically active bacteria will continuously emit blue green (490 nm) light without the need for the injection of an exogenous substrate12. Photons emitted by lux-tagged bacteria can be captured using highly sensitive, cooled charge-coupled device (CCD) cameras.
The use of bioluminescent bacteria in a model for UTI allows for the longitudinal, non-invasive quantification of the bacterial load, omitting the need for sacrificing animals at fixed time points during the follow-up for CFU determination. Despite the wide range of possibilities, accumulating evidence for the robustness of this BLI technique in other fields and its advantages over classic models of UTI, it has not been widely implemented in the UTI research. The protocol presented here provides a detailed step-by-step guide and highlights the advantages of BLI for all future UTI research.
Protocol
All animal experiments were conducted in accordance with the European Union Community Council guidelines and were approved by the Animal Ethics Committee of KU Leuven (P158/2018).
1. Culturing bacteria (adapted from7,13,14)
- Preparation
- Choose a luminescent UPEC strain that best fits the experimental needs.
NOTE: Here, the clinical cystitis isolate, UTI89 (E. coli), was selected because of its uropathogenic capacity in both humans and rodents, and its common use in murine UTI models5,7,15. The bioluminescent isogenic strain carrying the complete lux operon (luxCDABE) is further referred to as UTI89-lux. This operon also carries kanamycin sulfate (Km) resistance genes. Therefore, Km can be used to select bioluminescent colonies13. - Make a correlation curve for the CFU and optical density at 600 nanometers (OD 600 nm) for the chosen bacterial strain7.
- To do this, culture bacteria (using the protocol below) and make 8 different dilutions in phosphate buffered saline (PBS). Measure the OD at 600 nm for all these dilutions and plate them on Luria-Bertani (LB) plates to determine the CFU.
- Plot the OD 600 nm values and the CFU values to determine the correlation and to obtain a standard curve.
NOTE: For future experiments, measure the OD at 600 nm and use this standard curve to get an instant estimation of the CFU in a bacterial solution.
CAUTION: UPEC are pathogenic bacteria, ensure rooms are properly equipped and use personal protective equipment.
- Choose a luminescent UPEC strain that best fits the experimental needs.
- Three days before the instillation
- Obtain single colonies by streaking out the glycerol stock of bacteria, using an inoculation loop, on LB plates supplemented with 50 µg/mL Km and culture overnight at 37 °C. Seal these plates with a paraffin film to store at 4 °C up to 1 week.
- Two days before the instillation
- Fill a sterile 14 mL polystyrene round bottom tube with dual-position snap caps with 5 mL of LB broth supplemented with 50 µg/mL Km. Pick a single bacterial colony with an inoculation loop and add this to the LB broth. Vortex for 10 s to ensure proper mixing.
- Culture statically with the snap cap in the open position at 37 °C overnight.
NOTE: Static growth of E. coli promotes the expression of type 1-pili, which are critical for the adherence to and invasion of urinary epithelial cells. - Prepare a sterile Erlenmeyer or culture flask.
- One day before the instillation
- After the incubation, vortex the tube for 10 s to ensure proper homogenization of the bacterial culture. Make a sub-culture in the Erlenmeyer by adding 25 µL of the bacterial suspension to 25 mL of fresh LB medium without antibiotics.
- Close the Erlenmeyer and culture statically at 37 °C overnight.
- On the day of instillation
- Vortex the Erlenmeyer for 10 s to ensure proper mixing.
- Pour the culture from the Erlenmeyer into a 50 mL culture tube and centrifuge at 3,000 x g and 4 °C for 5 min. Decant the supernatant and resuspend the bacterial pellet in 10 mL of sterile PBS and vortex again for 10 s.
- Choose the experimental concentration of the inoculum (CFU) and use the standard curve obtained in step 1.1.2 to determine the corresponding OD 600 nm.
- Add 1 mL of the resuspended bacterial culture into 9 mL of sterile PBS and check the OD at 600 nm. Adjust further by adding either sterile PBS or concentrated bacterial solution, until the desired OD 600 nm is reached7.
NOTE: For example, for UTI89-lux adjust the culture by adding PBS until the OD 600 nm reaches 0.45, corresponding to 2 x 107 CFU/50 µL. To verify the number of viable CFUs, make 10-fold serial dilutions and plate 50 µL of the fifth and sixth 10-fold dilutions on LB plates in triplicate to obtain less than 300 colonies. Count the obtained colonies the next day after overnight incubation at 37 °C. OD 600 nm gives an instant read-out but use CFU as quality control.
2. Inoculation of the animals (adapted from7,16)
- Preparation of the animals
- Choose the desired mouse strain(s) depending on the research question and availability of knock-out lines, experimental details, and differences in UTI susceptibility5,17. Keep in mind that transurethral catheterization is easier in female mice. Do not use animals younger than 8 weeks, as they are immunologically immature. Here, 12-week-old female C57Bl/6J mice were used.
- Order animals well in advance and let them acclimate, ideally for 7 days. Group house animals in individually ventilated cages, under standard 12 h light/dark conditions.
- Shave the abdominal region of the animals to limit the loss of signal. Do not use hair removal cream, as it can burn the skin of the animals quickly. Restrain the animal by tightly holding the scruff and hind limbs with the non-dominant hand while shaving with the dominant hand. Alternatively, shave under isoflurane anesthesia.
NOTE: Animals were shaved 2 days prior to imaging, considering the animals will groom the shaved area even further. - Provide water and standard food ad libitum throughout the experiment. However, deprive animals from water 2 h prior to instillation to minimize the bladder volume during instillation.
- Mount a sterile 24 G angiocatheter tip on a 100 µL syringe and fill the syringe with the bacterial solution prepared in step 1.5.3.
NOTE: To determine the background luminescence, obtain a baseline image prior to the instillation (see step 3, below).
- Instillation of the animals
- Place the animals in an induction chamber and anesthetize them using inhalation of isoflurane with pure oxygen as a carrier gas (induction at 3% and maintenance at 1.5%).
- Place one animal on a working surface in the supine position and maintain a stable isoflurane anesthesia using a nose cone during the instillation. Apply the eye-ointment.
- Expel the residual urine by applying gentle compression and making circular movements on the suprapubic region. Clean the lower abdomen with 70% ethanol prior to instillation.
- Lubricate the catheter tip with normal saline. Put the index finger of the non-dominant hand on the abdomen and push it gently upwards. Start the catheterization of the urethra in a 90° angle (vertically) and once resistance is encountered, tilt it horizontally before inserting it further (0.5 cm).
NOTE: Never forcefully push the catheter, as this will cause harm to the urethra. Gentle turning motions can be helpful in catheter insertion. On the other hand, a lack of resistance usually indicates erroneous insertion into the vagina. - Perform a slow (5 µL/s) instillation of 50 µL of the bacterial inoculum (2 x 107 CFU).
NOTE: Higher volumes or faster instillation might cause reflux to the kidneys. During practice of the technique, blue ink can be used to evaluate reflux. - After the instillation, keep the syringe and catheter in place for a few more seconds and then slowly retract to prevent leakage. Record any irregularities such as a high amount of leakage or a bloody meatus and exclude animals, if necessary.
- Position the animal in supine position at the nose cone of the imaging chamber and repeat the preceding steps 2.1.1-2.1.6 for all the remaining animals. Use one catheter per experimental group. Ensure anesthesia is continued and minimize the time between the first and the last animal.
- If necessary, administer antibiotics or experimental drugs, prior to or after the imaging (step 3). For example, to administer enrofloxacin, add 40 µL of the enrofloxacin (100 mg/mL) solution to 3.96 mL of physiological saline to obtain a 1/100 dilution. Inject 100 µL/10 g subcutaneously at 9 am and 5 pm to administer 10 mg/kg of enrofloxacin twice daily for 3 days.
3. Bioluminescence imaging
- Preparation and selection of imaging parameters
- Open the BLI acquisition software (see Table of Materials) and click on Initialize in the imaging device (see Table of Materials) to test the camera and stage controller system and to cool the CCD camera to -90 °C.
NOTE: During this process, the door is locked, and the progress of the initialization can be followed on the control panel. A green light indicates that the temperature of -90 °C is reached. A warning will appear if imaging is attempted before completion of the initialization. - Ensure data is saved automatically: Click on acquisition Auto-save to and select the correct folder.
- Select Luminescence and Photograph. Check the default luminescence settings: Set excitation filter to Block and emission filter to Open.
- Set the exposure time to Auto when taking the first image, especially when expecting a dim signal to ensure an adequate number of photon counts. For in vivo measurements and bright signals, set the Exposure Time to ~30 s. If the image is saturated, a warning will appear. If this happens, reduce the exposure time.
- Select the medium Binning, F/stop 1 and choose the correct field of view (FOV) (D for 5 animals).
- Set the subject height to 1 cm when imaging mice.
- Click on Add three times in the Acquisition Control Panel to obtain a sequence of three images, as technical replicates.
- Open the BLI acquisition software (see Table of Materials) and click on Initialize in the imaging device (see Table of Materials) to test the camera and stage controller system and to cool the CCD camera to -90 °C.
- Imaging
- Place mice in the imaging chamber in the supine position and use the manifold nose cone to maintain anesthesia (isoflurane 1.5%) throughout the experiment. Image up to 5 mice simultaneously and separate the animals using the light baffle to prevent reflection.
- Close the door and click on Acquire to start the imaging sequence.
- Fill in detailed information about the experiment (wild type and knockout animals, treatments, day of imaging, etc.). The imaging settings, such as exposure time, are saved automatically.
- Remove mice from the imaging chamber and return them to their cage. Check for the full recovery after anesthesia. Within minutes, the animals should be fully awake and explorative. Do not provide analgesia as this might interfere with the UTI course18.
- Return the cages to the ventilated racks until the next imaging cycle. After completion of the experiment, euthanize the animals by CO2 asphyxiation or cervical dislocation. Do this before the recovery of isoflurane anesthesia to minimize distress.
- Analysis of the images
- Start the imaging software and load the experimental file by clicking on 열람.
- Use the Tool Palette to adjust the color scale of the image. Standardize the visual aspect of the results by using the same settings for all the images, i.e., use a logarithmic scale with a minimum of 104 to a maximum of 107 photons. These adjustments do not affect the raw data but only the graphical presentation.
- Use the ROI tools to draw a region of interest (ROI) on the image. Ensure the ROI is large enough to cover the complete area and use the same dimensions for all the images. Here, a square ROI of 3.5 cm x 4.5 cm was placed over the lower abdomen.
- Click on ROI measurement to quantify the light intensity (counts or total photon flux). Export these data and use the average of the technical replicates.
NOTE: Counts represent the number of photons detected by the camera and should only be used for the image quality control. To report data, use total photon flux. Total photon flux is more physiologically relevant as it represents the light emitted from the surface and it is a calibrated unit, which is corrected for the exposure time (per second).
Representative Results
In vivo BLI correlates with CFU of the inoculum at time of instillation.
To evaluate the detection limit of BLI in vivo and the correlation with CFU of the inoculum, mice were infected with different concentrations of UTI89-lux and PBS as a negative control. Before instillation, uninfected animals were scanned to determine the background luminescence. Subsequent images were obtained immediately post-instillation (Figure 1A). After the instillation of UTI89-lux, bioluminescence was robustly detectable above 2 x 104 CFU and a linear correlation between the CFU of the inoculum and the bioluminescence was established (Figure 1B). In contrast, total photon flux was comparable to background signals for animals instilled with PBS and at the lowest bacterial concentration of 2 x 103 CFU.
BLI as a tool for longitudinal follow-up during treatment
To determine whether the curative effect of a validated antibiotic treatment can be demonstrated using longitudinal BLI, 20 animals were infected with an inoculum of 2 x 107 CFU/50 µL, and 10 of these animals were treated with enrofloxacin 10 mg/kg subcutaneously twice daily during the first 3 days post-infection. Both the natural evolution in the infected but untreated control group and the effect of antibiotic treatment on the infection kinetics in the treated group could be visualized in detail (Figure 2). An immediate decrease in bacterial load, as measured by total photon flux, was seen after the first dose of enrofloxacin. None of the treated animals had a subsequent rise in bacterial load. The inter-subject variability in the infected but untreated control group was higher and the decrease in bacterial load was slower. At day 10, 8 out of 10 animals of the control group had returned to a BLI signal comparable to their pre-instillation background. The overall bacterial load for each animal was calculated using the area under the curve (AUC) of the log-transformed total photon flux. A significant difference was observed between animals treated with enrofloxacin and untreated animals over the time course of 10 days. These results demonstrate that BLI is a powerful tool to evaluate the effect of a potential treatment on the disease course of a UTI.
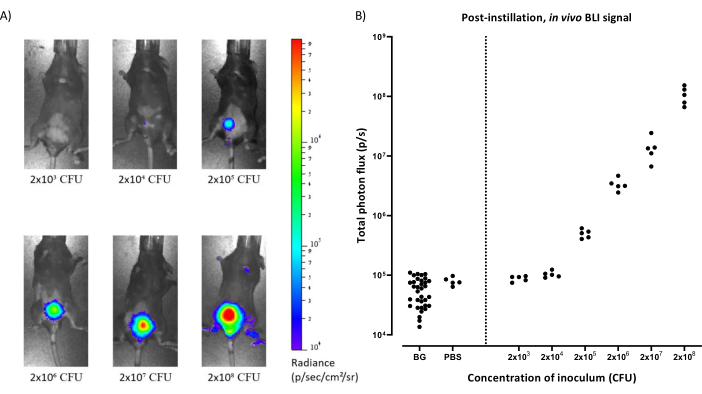
Figure 1: In vivo BLI correlates with CFU of the inoculum at the time of instillation. (A) Representative images obtained with in vivo BLI of mice infected with increasing concentrations of inoculum (2 x 103-2 x 108 CFU) imaged immediately after bacterial instillation in the bladder. CFU = Colony Forming Units.(B) Quantification of bioluminescence emitted over the bladder region (total photon flux) compared to the concentration of the inoculum (CFU). BLI signal was robustly detected above 2 x 104 CFU (p = 0.0002 compared to background and p = 0.015 compared to the PBS control, unpaired t-test). Pearson correlation for the inoculi ranging from 2 x 103-2 x 108 CFU was r = 0.9995 (p < 0.0001). BG = background, PBS = Phosphate Buffered Saline, CFU = Colony Forming Units. Please click here to view a larger version of this figure.
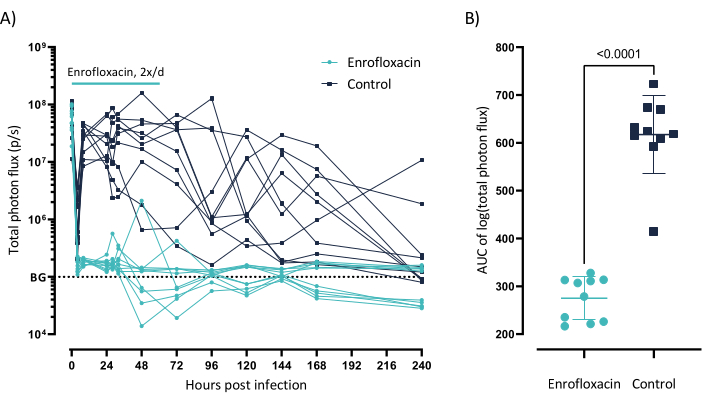
Figure 2: BLI as a tool for longitudinal follow-up during treatment. (A) Individual traces of the total photon flux for animals infected with 2 x 107 CFU of UTI89-lux and treated with enrofloxacin 10 mg/kg twice daily for 3 days (blue) versus an untreated control group (black), n = 10 per group. BG = background. (B) Analysis of traces from animals shown in panel A, reporting the AUC of log-transformed total photon flux for both the enrofloxacin (blue) and control group (black). N = 10 per group, mean ± SD, unpaired t-test p < 0.0001. AUC = Area under the curve. Please click here to view a larger version of this figure.
Discussion
Advantages of BLI compared to CFU counts
Longitudinal data
A major disadvantage of the traditional method of counting CFU to quantify microbial burden is the requirement of post-mortem organ homogenates, providing only one cross-sectional data point per animal. Conversely, BLI enables non-invasive longitudinal follow-up of infected animals. The animals can be imaged 2 to 3 times a day, providing detailed insight into the kinetics of the infection. Additionally, repeated measures of the same animal drastically reduce the number of animals needed for adequately powered experiments. Furthermore, researchers can focus on the inter-individual variability and prior to examining or treating animals with a novel agent, they can select animals with prespecified bacterial loads by using the real-time data. On the other hand, using the AUC of an experiment allows researchers to focus on the overall bacterial load and not on a single cross-sectional value. The added value of longitudinal and real-time data in the UTI setting cannot be overestimated.
Identification of dissemination of the infection
The spatial resolution of the images obtained with BLI is adequate to identify other sites that are colonized by lux- or FLuc-tagged pathogens19. For UTI, ascending infections to the kidneys or dissemination to the blood are highly relevant findings. In this experimental setting, any spread of infections induced by UTI89-lux beyond the lower urinary tract was not observed. This was expected considering this strain predominantly causes cystitis.
Data reproducibility and comparison
The reproducibility of experiments is higher when using BLI compared to CFU counts. Classically, triplicates of serial 10-fold dilutions of homogenates are plated and only those plates with 30 to 300 CFU are then counted to calculate the total number of CFU. This method is cumbersome, prone to high variability, results in many futile plates, and is extremely prone to human errors. Furthermore, there is no consensus on how to report the CFU, namely, per gram bladder tissue, per bladder, or per 1 mL homogenate. This renders the comparison of the results of different groups extremely difficult and could be ameliorated by the use of BLI and total photon flux.
Considerations and limitations of the method
Besides the classical considerations related to BLI in vivo such as absorption of photons by fur or hemoglobin, there are some considerations to be made when correlating BLI and CFU counts at the time of sacrifice.
First, discrepancies between both methods at the time of sacrifice are encountered20, especially when antibiotic therapies are used. Therefore, the correlation between BLI and CFU counts at the time of sacrifice can be suboptimal and is not necessarily relevant. For BLI, the total photon flux can be lower than expected, because challenged bacteria can redirect their metabolism toward recovery and repair processes, rather than light emission. Additionally, the BLI measurement captures a snapshot of the bacteria and their metabolism in real-time in vivo, resulting in lower BLI signals if bacteria are in a dormant or elongated state at the time of imaging. On the other hand, for CFU determination, drug-challenged bacteria react in an all-or-nothing manner after being removed from their infectious habitat (i.e., a biofilm) and plated. Once plated onto agar, the challenged bacteria may be irretrievably injured and unable to develop into observable colonies, resulting in lower CFU counts21. Additionally, challenged bacteria can enter a viable but non culturable state (VBNC). They remain metabolically active and viable but are no longer culturable on standard laboratory media. Bacteria that enter the VBNC state or that are organized in biofilms can be detected easily with BLI, while enumeration of CFU requires additional processing21,22,23. In conclusion, bioluminescence and viable counts measure different aspects of cell physiology and neither should be regarded as the definitive indicator of cell viability21.
Moreover, the definition of bacterial load as measured by BLI or CFU counts is different due to differences in sample processing. When using CFU counts, only bacteria that reside within the bladder wall are included in the homogenate and counted. By contrast, BLI also captures photons emitted by bacteria in the urine and the urethra. In our opinion, the latter is a physiologically more correct representation of the total bacterial load as it includes all anatomically relevant compartments (bladder, urine, and urethra).
In contrast to CFU counts, BLI requires the generation of genetically altered luminescent bacteria. Depending on the insertion site, it is plausible that the introduction of the lux operon in the genome can alter the virulence potential of the bacterial strain. Therefore, when developing a new bioluminescent strain, its virulence and growth characteristics should be compared to the parental strain10,13. However, recent advances in genome editing allow for efficient and scarless tagging of all major bacterial pathogens, thereby limiting the impact on virulence or treatment responses. Moreover, genetic engineering opens possibilities toward conditional expression of bioluminescence reporter genes and, therefore, in vivo measuring of relevant bacterial subpopulations, gene expression, etc24,25.
Finally, access to the advanced imaging device and acquisition software for in vivo BLI might be a limiting factor compared to the basic equipment required for CFU enumeration. However, collaborations with imaging cores can minimize the expenses and investments necessary for BLI.
Considerations specific to the implementation of BLI in an in vivo UTI model
Animal model
The advantages of a murine UTI model are three-fold: It allows monitoring of disease progression in a mammalian urinary tract, the animals and their maintenance are relatively inexpensive, and mutant lines are available. On the other hand, induction of the infection usually entails transurethral catheterization and subsequent instillation of a high dosage of bacteria directly into the bladder lumen, thereby bypassing the natural process of ascension via the urethra5.
Animal housing
Animals can be group housed during the experiment. We have examined the occurrence of transmission between animals of the same cage, by introducing sentinel animals into cages with infected animals. None of the sentinel animals had a detectable bacterial load, measured with both CFU and BLI.
Bladder volume
When combining the murine UTI model with BLI, researchers should keep the unique anatomical properties of the bladder in mind. The bladder is a hollow organ that contains a varying volume of urine. During UTI, the presence of bacteria-infested urine could theoretically influence the total photon count depending on the degree of bladder filling. However, standardization of the bladder volume is impractical and could intervene with the experimental design. Furthermore, in our experience, differences in bladder volume do not result in statistically significant differences in total photon flux.
Sample temperature
In vitro BLI (or as a simpler and cheaper alternative, measuring bioluminescence in a luminometer) can also be used to determine bacterial load on homogenates, urine, or bacterial suspensions containing UTI89-lux. However, the temperature of the sample is important as the emission of photons results from an enzymatic reaction and thus requires metabolic activity and the presence of oxygen. Drastic changes in temperature during organ harvesting should be avoided by allowing the sample to calibrate on the heated (37 °C) stage 5 min prior to imaging.
Potential applications and importance
In summary, the cumbersome method of enumeration of CFU was limiting the reproducibility and efficacy of research in the UTI field. The experimental setup with BLI will advance in vivo UTI research on bladder physiology, UTI pathogenesis, and susceptibility by enabling longitudinal follow-up, while reducing the number of animals needed for these types of studies.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This work was supported by grants from the Research Foundation – Flanders (FWO Vlaanderen; G0A6113N), the Research Council of KU Leuven (C1-TRPLe; T.V. and W.E.) and the VIB (to T.V.). W.E. is a senior clinical researcher of the Research Foundation – Flanders (FWO Vlaanderen). The strain UTI89-lux was a generous gift from Prof. Seed's laboratory13.
Materials
| 96-well Black Flat Bottom Polystyrene Plate | Corning | 3925 | for in vitro imaging |
| Aesculap ISIS | Aesculap | GT421 | hair trimmer, with GT608 cap |
| Anesthesia vaporizer | Harvard apparatus limited | N/A | https://www.harvardapparatus.com/harvard-apparatus-anesthetic-vaporizers.html |
| Baytril 100 mg/mL | Bayer | N/A | Enrofloxacin |
| BD Insyte Autoguard 24 GA | BD | 382912 | Yellow angiocatheter, use sterile plastic tip for instillation |
| C57Bl/6J mice | Janvier | N/A | |
| Centrifuge 5804R | Eppendorf | EP022628146 | |
| Dropsense 16 | Unchained Labs | Trinean | to measure OD 600nm |
| Dulbecco's Phosphate Buffered Saline, Gibco | ThermoFisher Scientific | REF 14040-083 | |
| Ethanol 70% denaturated 5L | VWR international | 85825360 | |
| Falcon 14ml Round Bottom Polystyrene Tube, Snap-Cap | Corning | 352057 | |
| Falcon 50ml cellstart | Greiner | 227285 | |
| Hamilton GASTIGHT syringe, PTFE luer lock, 100 µL | Sigma-Aldrich | 26203 | to ensure slow bacterial instillation of 50 µL |
| Inoculation loop | Roth | 6174.1 | holder: Art. No. 6189.1 |
| Iso-Vet 1000mg/g | Dechra Veterinary products | N/A | Isoflurane |
| IVIS Spectrum In Vivo Imaging System | PerkinElmer | REF 124262 | imaging device |
| Kanamycine solution 50 mg/mL | Sigma-Aldrich | CAS 25389-94-0 | |
| Living Imaging Software | PerkinElmer | N/A | BLI acquisition software, version 4.7.3 |
| Luria Bertani Broth | Sigma-Aldrich | REF L3022 | alternatively can be made |
| Luria Bertani Broth with agar | Sigma-Aldrich | REF L2897 | alternatively can be made |
| Petri dish Sterilin 90mm | ThermoFisher Scientific | 101VR20 | to fill with LB agar supplemented with Km |
| Pyrex Culture flask 250 mL | Sigma-Aldrich | SLW1141/08-20EA | |
| Slide 200 Trinean | Unchained Labs | 701-2007 | to measure OD 600nm |
| UTI89-lux | N/A | N/A | Generous gift from Prof. Seed |
| Vortex | VWR international | 444-1372 |
References
- Foxman, B. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. American Journal of Medicine. 113 (1), 5-13 (2002).
- O’Brien, V. P., Hannan, T. J., Nielsen, H. V., Hultgren, S. J. Drug and vaccine development for the treatment and prevention of urinary tract infections. Microbiology Spectrum. 4 (1), 1128 (2016).
- Nielubowicz, G. R., Mobley, H. L. Host-pathogen interactions in urinary tract infection. Nature Reviews Urology. 7 (8), 430-441 (2010).
- Foxman, B. The epidemiology of urinary tract infection. Nature Reviews Urology. 7 (12), 653-660 (2010).
- Carey, A. J., et al. Urinary tract infection of mice to model human disease: Practicalities, implications and limitations. Crititical Reviews in Microbiology. 42 (5), 780-799 (2016).
- Barber, A. E., Norton, J. P., Wiles, T. J., Mulvey, M. A. Strengths and limitations of model systems for the study of urinary tract infections and related pathologies. Microbiology and Molecular Biology Reviews. 80 (2), 351-367 (2016).
- Hung, C. S., Dodson, K. W., Hultgren, S. J. A murine model of urinary tract infection. Nature Protocols. 4 (8), 1230-1243 (2009).
- Contag, C. H., et al. Photonic detection of bacterial pathogens in living hosts. Molecular Microbiology. 18 (4), 593-603 (1995).
- Contag, P. R., Olomu, I. N., Stevenson, D. K., Contag, C. H. Bioluminescent indicators in living mammals. Nature Medicine. 4 (2), 245-247 (1998).
- Doyle, T. C., Burns, S. M., Contag, C. H. In vivo bioluminescence imaging for integrated studies of infection. Cellular Microbiology. 6 (4), 303-317 (2004).
- Hutchens, M., Luker, G. D. Applications of bioluminescence imaging to the study of infectious diseases. Cellular Microbiology. 9 (10), 2315-2322 (2007).
- Avci, P., et al. In-vivo monitoring of infectious diseases in living animals using bioluminescence imaging. Virulence. 9 (1), 28-63 (2018).
- Balsara, Z. R., et al. Enhanced susceptibility to urinary tract infection in the spinal cord-injured host with neurogenic bladder. Infection and Immunity. 81 (8), 3018-3026 (2013).
- Huang, Y. Y., et al. Antimicrobial photodynamic therapy mediated by methylene blue and potassium iodide to treat urinary tract infection in a female rat model. Scientific Reports. 8 (1), 7257 (2018).
- Mulvey, M. A., Schilling, J. D., Hultgren, S. J. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infection and Immunity. 69 (7), 4572-4579 (2001).
- Hannan, T. J., Hunstad, D. A. A murine model for E. coli urinary tract infection. Methods in Molecular Biology. 1333, 83-100 (2016).
- Hopkins, W. J., Gendron-Fitzpatrick, A., Balish, E., Uehling, D. T. Time course and host responses to Escherichia coli urinary tract infection in genetically distinct mouse strains. American Society for Microbiology. 66 (6), 2798 (1998).
- Zhang, Y., et al. Efficacy of Nonsteroidal Anti-inflammatory Drugs for Treatment of Uncomplicated Lower Urinary Tract Infections in Women: A Meta-analysis. Infectious Microbes & Diseases. 2 (2), 77-82 (2020).
- Vanherp, L., et al. Sensitive bioluminescence imaging of fungal dissemination to the brain in mouse models of cryptococcosis. Disease Models & Mechanisms. 12 (6), 039123 (2019).
- Keyaerts, M., Caveliers, V., Lahoutte, T. Bioluminescence imaging: looking beyond the light. Trends in Molecular Medicine. 18 (3), 164-172 (2012).
- Marques, C. N., Salisbury, V. C., Greenman, J., Bowker, K. E., Nelson, S. M. Discrepancy between viable counts and light output as viability measurements, following ciprofloxacin challenge of self-bioluminescent Pseudomonas aeruginosa biofilms. Journal of Antimicrobial Chemotherapy. 56 (4), 665-671 (2005).
- Vande Velde, G., Kucharikova, S., Van Dijck, P., Himmelreich, U. Bioluminescence imaging increases in vivo screening efficiency for antifungal activity against device-associated Candida albicans biofilms. International Journal of Antimicrobial Agents. 52 (1), 42-51 (2018).
- Oliver, J. D. Recent findings on the viable but nonculturable state in pathogenic bacteria. FEMS Microbiology Reviews. 34 (4), 415-425 (2010).
- Kucharikova, S., Van de Velde, G., Himmelreich, U., Van Dijck, P. Candida albicans biofilm development on medically-relevant foreign bodies in a mouse subcutaneous model followed by bioluminescence imaging. Journal of Visualized Experiments: JoVE. (95), e52239 (2015).
- Van de Velde, G., Kucharikova, S., Schrevens, S., Himmelreich, U., Van Dijck, P. Towards non-invasive monitoring of pathogen-host interactions during Candida albicans biofilm formation using in vivo bioluminescence. Cellular Microbiology. 16 (1), 115-130 (2014).

