3-D Time-Lapse Imaging of Cell Wall Dynamics Using Calcofluor in the Moss Physcomitrium patens
Summary
This manuscript presents a detailed protocol to image the 3-D cell wall dynamics of living moss tissue, allowing the visualization of the detachment of cell walls in ggb mutants and thickening cell wall patterns in the wild type during development over a long period.
Abstract
Time-lapse imaging with fluorescence microscopy allows observation of the dynamic changes of growth and development at cellular and subcellular levels. In general, for observations over a long period, the technique requires transformation of a fluorescent protein; however, for most systems, genetic transformation is either time-consuming or technically unavailable. This manuscript presents a protocol for 3-D time-lapse imaging of cell wall dynamics over a 3 day period using calcofluor dye (which stains cellulose in the plant cell wall), developed in the moss Physcomitrium patens. The calcofluor dye signal from the cell wall is stable and can last for 1 week without obvious decay. Using this method, it has been shown that the detachment of cells in ggb mutants (in which the protein geranylgeranyltransferase-I beta subunit is knocked out) is caused by unregulated cell expansion and cell wall integrity defects. Moreover, the patterns of calcofluor staining change over time; less intensely stained regions correlate with the future cell expansion/branching sites in the wild type. This method can be applied to many other systems that contain cell walls and that can be stained by calcofluor.
Introduction
Plant cell walls undergo dynamic changes during cell expansion and development1,2,3. Maintaining cell wall integrity is critical for plant cell adhesion during growth and development, as well as for the response to environmental signals. Although visualizing cell wall dynamics of living cells over a long period of time is critical to understanding how cell adhesion is maintained during development and adaptation to environmental changes, current methods for directly observing cell wall dynamics are still challenging.
Time-lapse imaging of cellular changes can provide informative developmental dynamics of an organism using a high-resolution fluorescence microscope4,5,6,7. While time-lapse 3-D imaging has a great deal of potential for studying dynamic changes of cell shape during growth and development, the technique normally requires transformation of a fluorescent protein4,5,6,7. However, for most systems, genetic transformations are either time-consuming or technically challenging. As an alternative, fluorescent dyes that attach to cellular components have long been available. The fluorescent dyes can emit fluorescent light after irradiation with light of a certain wavelength. Common examples are Edu, DAPI, PI, FM4-64, and calcofluor white8,9,10. One major drawback, however, is that these dyes can typically only be used in fixed tissue or for short experiments, in part due to the harm they cause to the cell8,9,10.
With the protocol presented here, calcofluor signals are stable when calcofluor white is mixed within the medium during time-lapse experiments in the moss P. patens. Using this method, the detachment of cells in ggb mutants using 3-D time-lapse imaging was observed over a 3 day period3 (Figure 1). This method can be applied to many other systems that contain cell walls and that can be stained by calcofluor.
Protocol
NOTE: See the Table of Materials for the list of materials and equipment and Table 1 for the list of solutions to be used in this protocol.
1. Preparation of plants for glass bottom dishes
- Grow the moss protonemal tissue in BCDAT agar medium in a 13 cm Petri dish under constant white light (~50 µmol m-2s-1) for 7 days at 25 °C in the growth chamber.
- Filter-sterilize the calcofluor white and store at 4 °C until use. The reagent is stable for several months.
- Disperse the 7-day-old moss tissue into a 1.5 mL microtube containing 20 µL of sterilized water. Ensure the moss protonemal tissue is sufficiently dispersed as small pieces in the water. For wild type (WT), use forceps to separate the protonemata; for the ggb mutant, use a P-1000 pipette.
- Add 10 µL of the sterilized calcofluor white into the 1.5 mL microtube and leave the microtube upright in the hood for 2 min for staining.
- Immediately after staining, mix the plants with 200 µL of cooled BCDATG, containing BCDAT medium supplemented with 0.5% (w/v) glucose and 0.4% (w/v) gellan gum, by pipetting five times with a P-1000 pipette.
NOTE: Ensure the BCDATG medium is cooled enough (just before the medium solidifies) to avoid stressing the plants. - Transfer the mixture containing the plants and the BCDATG medium into a 27 mm diameter glass base dish. Ensure that the volume of BCDATG for mixing the cells is no more than 200 µL and that the 200 µL of BCDATG with samples is evenly distributed on the surface of the 7 mm glass bottom dish to produce a thin layer of film, so that the samples are within the objective working distance during imaging.
- After solidifying for 2 min at room temperature, cover the thin layer with 3 mL of cooled BCDATG and let it set for 10 min to solidify again.
NOTE: Ensure that steps 1.2-1.5 are conducted in a sterile hood to avoid contamination. - On the bottom of the dish, draw nine lines each in parallel and perpendicular directions (Figure 2). Mark the rows with characters from a to h, and mark the columns with numbers from 1 to 8. Use upper, lower, right, middle, and left to mark the positions within each square. For example, if a plant is in a square in the second row and the sixth column and is positioned in the upper left part of the square, then this plant is given the name of "b6_upper_left".
- Preculture the glass bottom dish containing the plants under red light for 5 days at 25 °C in the growth chamber, and then under white light for 1-2 days before being subjected to time-lapse imaging every day for up to 3 days. For WT, preculture the plants in weak red light (0.5 µmol m-2s-1) from the above for 5 days to induce the phototropic response of the protonemata, so that the plants can attach to the bottom of the dishes11.
NOTE: The weak red light is provided by passing a white light through a 3 mm thick red acrylic plastic filter into light-proof boxes. For ggb mutants, preculturing in red light is optional, as the ggb mutants do not have a phototropic response3.
2. Imaging and 3-D reconstruction
- Place the glass bottom dish containing plants onto the stage of an inverted confocal microscope, with the glass bottom facing the objective. Use a confocal microscope for the visualization of calcofluor with a 20x objective lens. Take images with the 405 nm laser, the pinhole at 1.0, HV-gain at 100, offset-background adjustment at 0, Laser Power at 5%-7%, a scan size of 1,024 x 1,024, and a scan speed of 1/2 frame/s. Collect 17-30 z-series optical sections with a step size of 1.0 µm. Name the images using the code in step 1.6, such as "b6_upper_left-day0", and save.
- Select the DAPI channel in Acquisition and check the Z box in ND Acquisition. To set the lower and upper limits for the Z-stack, click the Top and Bottom buttons.
- To reconstruct the 3-D images, open the .nd2 file (Figure 3A; see Table of Materials) and click the Volume (Figure 3B).
3. Imaging the same plants at different time points
- After each imaging, put the glass base dish back into the growth chamber.
- Repeat from step 2.1 for imaging and 3-D reconstruction.
NOTE: To find the same plants, use the code mentioned in step 1.6 and give the name "b6_upper_left-day1", or "b6_upper_left-day2", according to the age of plants.
Representative Results
This method allows the observation of cell wall dynamics during development in wild type and ggb mutants (Figure 1). The results showed that regions with less thickening of the cell wall correlate with the cell expansion/branching sites, allowing for the prediction of expansion/branching sites in the wild type (Figure 1A). The surface of the cell walls in ggb mutants was torn apart during development due to uncontrolled cell expansion3 (Figure 1B). Moreover, the staining signal of the broken surface of ggb mutants (which is older cellulose) is stronger than the cell surface (which is younger cellulose) beneath the broken cell wall surface.
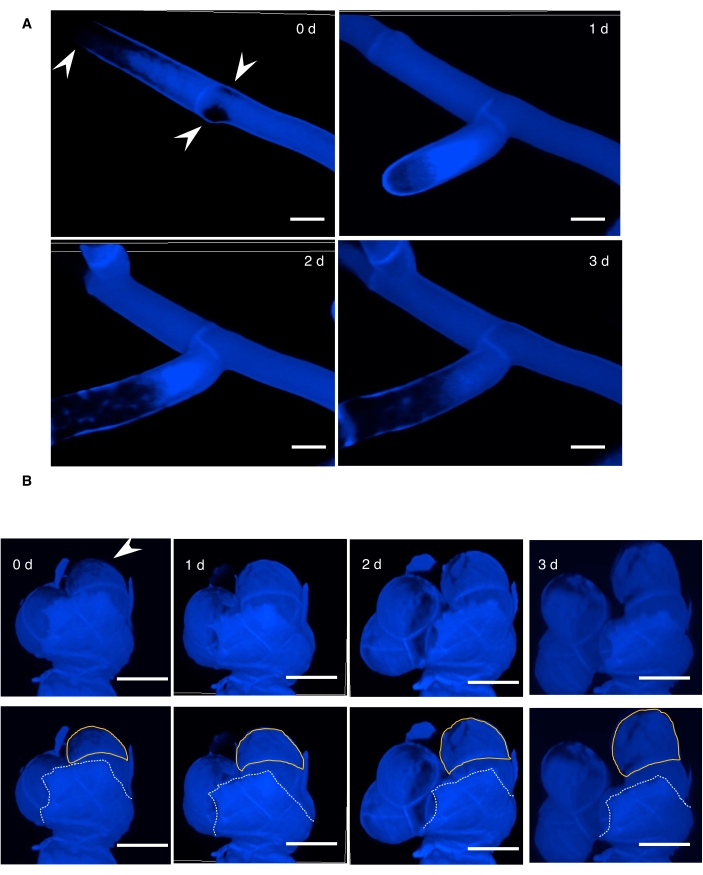
Figure 1: 3-D time series of moss development. (A,B) Dynamic changes in cross-walls were shown by calcofluor white staining (which stains cellulose) in WT (A) and ggb mutants (B). For ggb mutants (B), white dash lines below each image indicate the broken surface cell walls on the expanding cells, and orange lines indicate the cells beneath the cell wall surface in ggb. Panel B is reprinted with permission from3. Note that the differences in calcofluor white signal can be detected on the surface positions of the cells, and less dense staining regions (arrowheads) are correlated to the cell expansion/branching sites. Scale bars = 20 µm. Abbreviations: d = days; WT = wild type; ggb = geranylgeranyltransferase-I beta. Please click here to view a larger version of this figure.
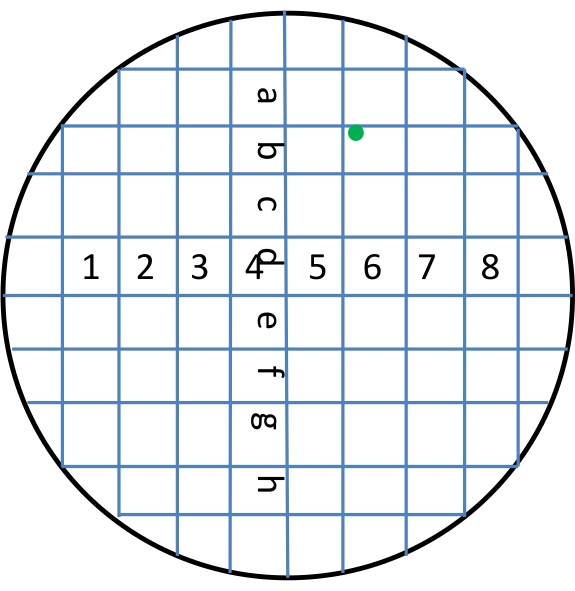
Figure 2: Marking location of plants. On the bottom of the dish, nine lines each were drawn in parallel and perpendicular directions to mark the location of squares. Characters a to h were used for marking rows, and numbers 1 to 8 were used for marking columns. Upper, lower, right, middle, and left were used for marking positions within each square. For example, if a plant (denoted by a green dot) was located in a square at the second row and the sixth column, and positioned in the upper left part, then the plant was named b6_upper_left. Please click here to view a larger version of this figure.
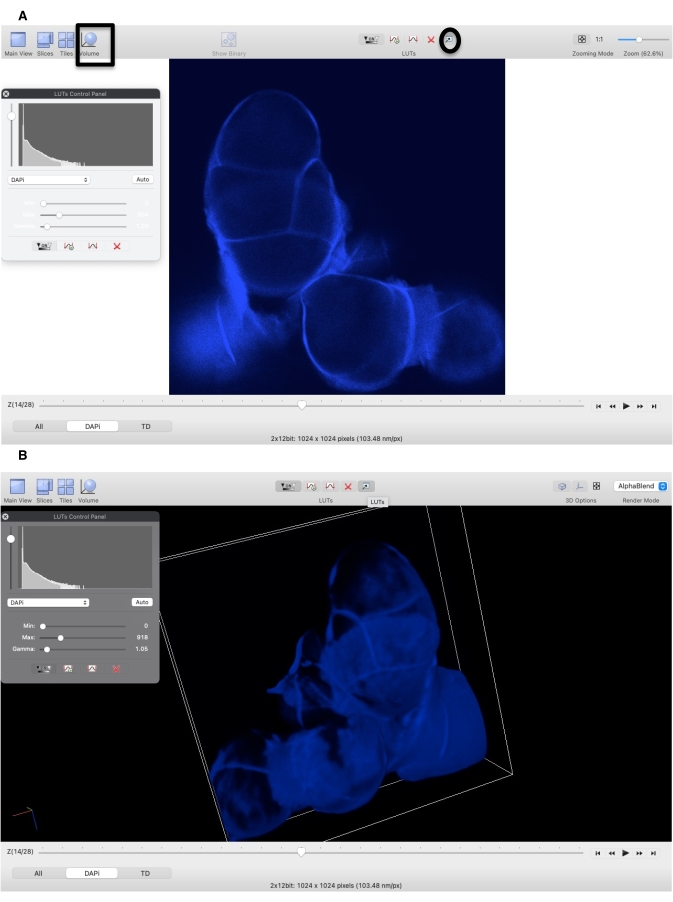
Figure 3: Reconstruction of 3-D images using a nd2 z-stack file. (A) Open an nd2 file in the NIS-elements Viewer. Click LUTs (circle) to adjust the contrast and brightness of images and click Volume (square) to reconstruct the 3-D image. (B) Reconstructed 3-D image from A. Please click here to view a larger version of this figure.
Table 1: List of solutions used in this protocol. Please click here to download this Table.
Discussion
Time-lapse 3-D reconstruction, or 4-D imaging, is a powerful tool for observing the dynamics of cellular morphology during developmental processes. In this protocol, by mixing the calcofluor white in the medium, the dynamics of 3-D cellular morphology can be observed in the moss P. patens. Using this method, we observed that the surface of cell walls in ggb mutants are torn apart during development3. Moreover, the reduced thickening of cell walls is correlated with the cell expansion/branching sites in wild type, allowing for prediction of the expansion/branching sites. Further, because calcofluor white can be used with other dyes, such as microspheres, additional information about cell expansion can be observed3.
There are two reasons that could explain how the calcofluor dye can bind for days to plants without affecting/damaging the molecular target. First, the low working concentration (10 µL in 200 µL of BCDATG medium, or 5%) may be sufficiently low for the moss protonemata to not be affected. Second, the calcofluor may stain the mature stage of cellulose, because the staining signal is much stronger in the basal part of wild type (which is older tissue) than the apical region of the apical cell (which is younger tissue), and the broken cell wall surface of ggb mutants (which is older cellulose) is stronger than the cells (which is younger cellulose) beneath the broken cell wall surface.
This protocol has been used for moss tissue, which is simple in structure. The current protocol may also be applied to other systems with simple structures, such as root hairs and trichomes in flowering plants. However, it remains to be seen if the calcofluor white staining works for other systems with more cell layers, such as stems. Moreover, because the calcofluor white is mixed with the medium, the tissue to be stained must be immersed within the medium. Therefore, optimization may be required in order to apply this protocol to other systems.
Disclosures
The authors have nothing to disclose.
Acknowledgements
The authors thank Dr. Soucy Patricia and Betty Nunn at the University of Louisville for assistance with the confocal microscope. This work was funded by the National Science Foundation (1456884 to M.P.R.) and by a National Science Foundation Cooperative Agreement (1849213 to M.P.R.).
Materials
| 3-mm-thick red plastic light filter | Mitsubishi | no.102 | |
| 27 mm diameter glass base dish | Iwaki | 3930-035 | |
| Agar | Sigma | A6924 | |
| Calcofluor white | Sigma | 18909-100ML-F | Calcofluor White M2R, 1 g/L and Evans blue, 0.5 g/L |
| Confocal microscope | Nikon | A1 |
NIS element software; .nd2 file in NIS-elements Viewer, download from https://www.microscope.healthcare.nikon. |
| Fluorescence microscope | Nikon | TE200 | Equipped with a DS-U3 camera; |
| Gellan gum | Nacali Tesque | 12389-96 | |
| Plant Growth Chambers | SANYO | Sanyo MLR-350H | |
| Sterilized syringe 0.22 μm filter | Millipore | SLGV033RS |
References
- Anderson, C. T., Kieber, J. J. Dynamic construction, perception, and remodeling of plant cell walls. Annual Review of Plant Biology. 71, 39-69 (2020).
- Vaahtera, L., Schulz, J., Hamann, T. Cell wall integrity maintenance during plant development and interaction with the environment. Naure Plants. 5 (9), 924-932 (2019).
- Bao, L., et al. The cellular function of ROP GTPase prenylation important for multicellularity in the moss Physcomitrium patens. Development. 149 (12), (2022).
- Colin, L., et al. Imaging the living plant cell: From probes to quantification. The Plant Cell. 34 (1), 247-272 (2022).
- Fang, Y., Spector, D. L. Live cell imaging of plants. Cold Spring Harbor Protocols. 2010 (2), (2010).
- Goh, T. Long-term live-cell imaging approaches to study lateral root formation in Arabidopsis thaliana. Microscopy. 68 (1), 4-12 (2019).
- Hamant, O., Das, P., Burian, A. Time-lapse imaging of developing shoot meristems using a confocal laser scanning microscope. Methods in Molecular Biology. 1992, 257-268 (2019).
- Rigal, A., Doyle, S. M., Robert, S. Live cell imaging of FM4-64, a tool for tracing the endocytic pathways in Arabidopsis root cells. Methods in Molecular Biology. 1242, 93-103 (2015).
- Yagi, N., et al. Advances in synthetic fluorescent probe labeling for live-cell imaging in plants. Plant & Cell Physiology. 62 (8), 1259-1268 (2021).
- Whitewoods, C. D., et al. CLAVATA was a genetic novelty for the morphological innovation of 3D growth in land plants. Current Biology. 28 (15), 2365-2376 (2018).
- Bao, L., et al. A PSTAIRE-type cyclin-dependent kinase controls light responses in land plants. Science Advances. 8 (4), 2116 (2022).

