Evaluating the Immune Response of a Nanoemulsion Adjuvant Vaccine Against Methicillin-Resistant Staphylococcus aureus (MRSA) Infection
Summary
The present protocol prepares and evaluates the physical properties, immune response, and in vivo protective effect of a novel nanoemulsion adjuvant vaccine.
Abstract
Nanoemulsion adjuvant vaccines have attracted extensive attention because of their small particle size, high thermal stability, and ability to induce validly immune responses. However, establishing a series of comprehensive protocols to evaluate the immune response of a novel nanoemulsion adjuvant vaccine is vital. Therefore, this article features a rigorous procedure to determine the physicochemical characteristics of a vaccine (by transmission electron microscopy [TEM], atomic force microscopy [AFM], and dynamic light scattering [DLS]), the stability of the vaccine antigen and system (by a high-speed centrifuge test, a thermodynamic stability test, SDS-PAGE, and western blot), and the specific immune response (IgG1, IgG2a, and IgG2b). Using this approach, researchers can evaluate accurately the protective effect of a novel nanoemulsion adjuvant vaccine in a lethal MRSA252 mouse model. With these protocols, the most promising nanoemulsion vaccine adjuvant in terms of effective adjuvant potential can be identified. In addition, the methods can help optimize novel vaccines for future development.
Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) is an opportunistic pathogen with one of the highest infection rates in an intensive care unit (ICU) wards1, cardiology departments, and burn departments worldwide. MRSA exhibits high rates of infection, mortality, and broad drug resistance, presenting great difficulties in clinical treatment2. In the Global Priority List of Antibiotic-Resistant Bacteria released by the World Health Organisation (WHO) in 2017, MRSA was listed in the most critical category3. A vaccine against MRSA infection is therefore urgently needed.
Aluminum adjuvant has been used for a long time, and the adjuvant auxiliary mechanism is relatively clear, safe, effective, and well tolerated4. Aluminum adjuvants are currently a widely used type of adjuvant. It is generally believed that antigens adsorbed on aluminum salt particles can improve the stability and enhance the ability of the injection site to uptake antigens, providing good absorption and slow release5. Currently, the main disadvantage of aluminum adjuvants is that they lack an adjuvant effect or exhibit only a weak adjuvant effect on some vaccine candidate antigens6. In addition, aluminum adjuvants induce IgE-mediated hypersensitivity reactions5. Therefore, it is necessary to develop novel adjuvants to stimulate a stronger immune response.
Nanoemulsion adjuvants are colloidal dispersion systems composed of oil, water, surfactants, and cosurfactants7. In addition, the adjuvants are thermodynamically stable and isotropic, can be autoclaved or stabilized by high-speed centrifugation, and can be formed spontaneously under mild preparation conditions. Several emulsion adjuvants (such as MF59, NB001-002 series, AS01-04 series, etc.) are currently on the market or in the clinical research stage, but their particle sizes are greater than 160 nm8. Therefore, the advantages of nanoscale (1-100 nm) medicinal preparations (i.e., large specific surface area, small particle size, surface effect, high surface energy, small size effect, and macro quantum tunneling effect) cannot be fully exploited. In the present protocol, a novel adjuvant based on nanoemulsion technology with a diameter size of 1-100 nm has been reported to exhibit good adjuvant activity9. We tested the recombination subunit vaccine antigen protein HI (α-hemolysin mutant [Hla] and Fe ion surface determining factor B [IsdB] subunit N2 active fragment fusion protein); a series of procedures were established to examine the physical properties and stability, evaluate its specific antibody response after intramuscular administration, and test the protective effect of the vaccine using a mouse systemic infection model.
Protocol
The animal experiments were conducted based on the manual on the use and care of experimental animals and were approved by the Laboratory Animal Welfare and Ethics Committee of the Third Military Medical University. Female Balb/c mice, 6-8 weeks old, were used for the present study. The animals were obtained from commercial sources (see Table of Materials).
1. Preparation of the MRSA HI antigen protein
- Obtain IsdB and Hla clones from commercial sources (see Table of Materials), perform polymerase chain reaction (PCR) amplification to amplify IsdB and Hla genes, and ligate their products to the PGEX-6P-2 plasmid (obtained commercially; see Table of Materials). Screen with the vector Xl/blue strain of Escherichia coli10.
- Transform the positive clone into E. coli BL21 competent cells (see Table of Materials) and amplify with shark culture10.
- Express the antigen and isolate after induction with isopropyl-β-D-1-mercaptogalactopyranoside and multimodal chromatography (MMC) isolate kits11(see Table of Materials).
- Characterize the antigen protein and purify it with a full-automation purifier11.
- Affinity-purify gutathione S-transferase (GST)-tagged proteins from cleared lysates using a commercially available affinity chromatography resin (see Table of Materials).
- Purify the recombinant proteins by MMC11.
- Remove the endotoxin by the Triton X-114 phase separation method11. Briefly, mix 1% Triton X-114 with recombinant protein eluate at 0 °C for 5 min to ensure a homogenous solution, and incubate at 37 °C for 5 min to allow two phases to form.
- Use the bacterial endotoxin test method11 with Turtle kits (see Table of Materials) to detect the endotoxin concentration. The endotoxin for protein antigen is 0.06 EU/µg, lower than the international standard (1.5 EU/µg)10.
- Perform an interference test to eliminate the possible influence of the test product on endotoxin detection11.
NOTE: According to the Appendix 2020 of Chinese pharmacopeia12, the endotoxin content should be less than 5 EU/dose. The sensitivity of Limulus lysate (λ0.25 EU/mL) and the maximum effective dilution ratio of 33.3 were calculated12 based on MVD = cL/λ, where L is the limit value of bacterial endotoxin for the test article, c is the concentration of the test article solution, and when L is expressed in EU/m, c is equal to 1.0 mg/mL. λ is the labeled sensitivity (EU/mL) of the horseshoe crab reagent in the gel method.
- Perform an interference test to eliminate the possible influence of the test product on endotoxin detection11.
- Recombine the antigen (48 kDa, determined by 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE]) at 1.5 mg/mL in sterile water without endotoxin or phosphate buffered saline (PBS) (sodium dichlorate method)11.
NOTE: The protein peak time was 13.843 min, the purity was 99.4% (High-performance liquid chromatography method), and the protein was stored at -70 °C10. Genomic DNA extracted from S. aureus strain MRSA252 (see Table of Materials) was used as the PCR template. The IsdB gene was amplified using the forward primer 59-CGCGGATCCATGAATGGCGAAGCAAAAGCAG
C-39 and the reverse primer 59-TTTTCCTTTTGCGG
CCGCCTATGTCATATCTTTATTAGATTCTTC-39. The Hla gene was amplified using the forward primer PHLAF (GCGGATCCGATTCTGATATTAATATTAAAACC) and the reverse primer PHLAR (TAAGCGGCCGCTTAT
CAATTTGTCATTTCTTC).
2. Preparation of the nanoemulsion vaccine
- According to the mass ratio of 1:6:3(w/w), add three kinds of excipients in sequence (isopropyl myristate, polyoxyethylene castor oil, and propylene glycol; see Table of Materials) to the HI antigen protein solution (10 mg/mL) and stir well. Then, add gradually sterilized pure water to reach a final volume of 10 mL and stir at 500 rpm and 25 °C for approximately 20 min.
NOTE: The nanoemulsion prepared in this protocol has a volume of 10 mL, and the masses of the three excipients are 0.34 g, 2 g, and 1 g, respectively. The protein solution here is the working solution, 10 times the concentration of the final solution. - Prepare the nanoemulsion adjuvant vaccine (1 mg/mL protein) by low-energy emulsification methods13 to obtain a clear and transparent liquid solution.
NOTE: The blank nanoemulsion (BNE) was prepared by similar methods, except that water was replaced with HI. The usual emulsification method involves heating the outer and inner phases to approximately 80 °C for emulsification and then stirring and cooling them gradually. In this process, a large amount of energy is consumed, and finally, the emulsion is obtained.
3. Physical characterization and stability of the nanoemulsion adjuvant vaccine
- Particle size, zeta potential, and polymer dispersion index characterization (PDI).
- Prepare 100 µL of the nanoemulsion adjuvant vaccine and dilute 50-fold with water for injection. Add 2 mL of sample for detection.
- Observe the particle sizes, zeta potential, and PDI at 25 °C using a Nano Zetasizer (ZS; see Table of Materials).
- Transmission electron microscopy (TEM)
- Dilute 10 µL of nanoemulsion vaccine 50-fold with water, place on a carbon-coated 100-mesh copper grid, and stain with 1% phosphotungstic acid (see Table of Materials) for 5 min at room temperature.
- Remove excess phosphotungstic acid solution with filter paper.
- Observe the morphology under a transmission electron microscope (see Table of Materials). Examine whether the finished products are nearly stable and circular within the field of view of the electron microscope and whether they exhibit good dispersion.
- Scanning electron microscopy (SEM)
- Dilute 10 µL of the nanoemulsion adjuvant vaccine 50-fold with water, drop 10 µL of prediluted samples on a silicon wafer, and place in a 37 °C thermostat-controlled incubator for 24 h.
- Spray the prepared platinum on the silicon for 40 s and cool to room temperature.
- Observe the morphological properties of the prepared samples under a high-resolution scanning electron microscope (see Table of Materials). Determine whether the samples are nearly stable, spherical, and well dispersed within the field of view under the electron microscope.
- Morphological stability
- Place 1 mL of nanoemulsion vaccine samples in microcentrifuge tubes, and evaluate the stability of fresh samples by centrifuging for 30 min at 13,000 x g at room temperature (25 °).
- Observe the appearance of these fresh samples and then perform a thermodynamic stability test of six temperature cycles (one cycle: store at 4 °C for 48 h, 25 °C for 48 h, and 37 °C for 48 h).
- After centrifugation, allow the samples to stand for 15 min to determine whether there is a Tyndall effect (whether the sample is clear and transparent under light) and whether demulsification has occurred (shown by clear stratification after demulsification).
- SDS-PAGE and western blot
- Shake the samples, mix a solution of 0.6 mL of absolute ethanol and 0.3 mL of vaccine, and centrifuge (13,000 x g, 30 min at room temperature, 25 °C).
- Transfer the supernatant of the solution to a clean tube and dissolve the precipitate with 0.3 mL of water. Obtain the supernatant and precipitate solutions (both blank nanoemulsion and vaccine) after treatment with absolute ethanol and distilled water solution, and perform the same 50-fold dilution.
- Mix 2.5 mL of separation gel A and 2.5 mL of separation gel B (see Table of Materials), add 50 µL of 10% ammonium persulfate, and quickly place the solution in the glass plate with a pipette. Mix 1 mL of concentrated gel A and 1 mL of concentrated gel B, add 20 µL of 10% ammonium persulfate into the glass plate, insert an appropriately sized comb, and wait for solidification.
- Add a total of 5 µL of 5x protein loading buffer solution to the diluted sample obtained in step 3.5.2, and boil the sample for 5 min.
- Add 10 µL of heated protein to the corresponding well, and add 5 µL of protein marker to the marker well.
- Connect the electrode, turn on the electrophoresis meter (see Table of Materials), and adjust the voltage to 300 V. Turn off the power when the electrophoresis reaches 1 cm away from the bottom of the PAGE rubber.
- Remove the gel and rinse with ddH2O. Add Coomassie bright blue G-250 staining solution (see Table of Materials) for 10 min. Pour out the staining solution, and rinse the gel twice with ddH2O.
- Add the commercially available decolorization solution and microwave the gel for 1 min. Shake the gel for 10 min, and repeatedly change the decolorization solution until the gel background is clear. Then, perform gel imaging (see Table of Materials).
NOTE: The western blot was preprocessed, as described in steps 3.5.1-3.5.6. - Soak three filter paper blocks with an electrophoresis transfer buffer. Soak the polyvinylidene fluoride (PVDF) membrane in methanol for 30 s and transfer with a semidry gel transfer instrument (two layers of filter paper, PVDF membrane, electrophoresis gel, and one layer of filter paper). Use a voltage of 23 V to transfer the membrane for 23 min.
- Remove the PVDF membrane and wash once with tris-buffered saline-Tween 20 (TBST) after transfer.
- Place the PVDF membrane in sealing solution (5% skimmed milk powder solution) and seal overnight at 4 °C.
- Remove the PVDF membrane and wash three times with TBST or 10 min each.
- Incubate with the primary antibody (from immunized mice with positive serum). Dilute the primary antibody11 (see Table of Materials) to be tested 500-fold (100-fold) with TBST. Place the blocked PVDF membrane in the primary antibody and incubate at 37 °C for 1 h.
- Remove the PVDF membrane and wash three times with TBST for 10 min each.
- Incubate with the secondary antibody (see Table of Materials). Dilute alkaline phosphatase (AP)-labeled goat anti-mouse secondary antibody 5,000-fold with TBST. Place the PVDF membrane in the secondary antibody and incubate at 37 °C for 40 min.
- Remove the PVDF membrane and wash four times with TBST for 10 min each.
- Submerge the PVDF membranes in a mixture (just enough to soak the membrane) of the AP substrate nitro blue tetrazole (NBT) and BCIP (5-bromo-4-chloro-3'-indolyphosphate p-toluidine salt) (see Table of Materials) for staining. A positive result is purple.
NOTE: The results must be a clear purple band, and the sample strip and the molecular weight of the antigen should be at the same labeled position.
4. Assessment of antibody immune response to this vaccine after intramuscular administration
NOTE: The mice were immunized by intramuscular injection of the nanoemulsion vaccine following a published report11. The mice were administered PBS as a negative control. At 1 week after three immunizations were completed, serum was collected from the mice11. The serum levels of IgG and subclasses of IgG1, IgG2a, and IgG2b were quantitatively determined by enzyme-linked immunosorbent assay (ELISA).
- Antigen coating: Dilute HI protein antigen to 10 µg/mL with coating solution and add to the ELISA plate at 100 µL/well. Incubate at 4 °C overnight.
NOTE: The coating solution is prepared by dissolving 1.6 g of anhydrous sodium carbonate and 2.9 g of sodium bicarbonate in 1 L of deionized water. - Sealing: Wash the plate (three cycles, each cycle 300 µL). Add 300 µL of the sealing solution to each well, and seal the wells for 2 h at 37 °C.
NOTE: The sealing solution is prepared by adding Tween-20 and bovine serum albumin (BSA) to PBS. The content of BSA in the PBST solution is 1%. - Serum dilution: Dilute the sample serum of mice 100 times with PBST (take 3 µL of serum and add it to 297 µL of PBST and vortex), and use 100 µL for each serum sample. Dilute the positive serum and negative control serum 100 times with PBST by adding 4 µL to 396 µL of PBST, then mixing by vortexing.
- Double ratio dilution: Add 200 µL of the above diluted experimental serum to the first row of the coated ELISA plate, and add 100 µL of PBST to all wells except the first and 12th rows. Perform a 1:2 dilution successively from the first row: adjust the pipette to 100 µL, transfer 100 µL from the first row to the second row, and mix with the pipette 10 times.
- Dilute the serum to Row 11 at multiple ratios and discard 100 µL of liquid.
- Add diluted negative and positive serum to the corresponding wells in Row 12 according to the sample template (see Table 1)-100 µL/well.
- Seal the ELISA plates with plastic wrap and incubate at 37 °C for 1 h.
- Dilute secondary antibody anti-mouse IgG-HRP (see Table of Materials) with PBST at 1:10,000.
- Wash the plate (three cycles, each cycle 300 µL). Then, add 100 µL of the diluted secondary antibody to each well, and incubate the plate at 37 °C for 40 min.
- Staining and termination: Wash the plate (five cycles, each cycle 300 µL). Add 3,3′,5,5′-tetramethylbenzidine (TMB) staining solution (100 µL; see Table of Materials) to each well. Place the plates at 37 °C for 10 min away from light, and then terminate the reaction immediately with 50 µL of 2 M H2SO4.
- Detect the optical density (OD450) value by an enzyme marker (read the OD450 value within 15 min after termination).
5. Evaluation of the in vivo effects of the novel adjuvant vaccines
NOTE: The protective effect of this nanoemulsion vaccine was evaluated in a commercially obtained lethal MRSA252 mouse model (see Table of Materials). According to the previous results of our research group14, 1 x 108 colony forming units (CFU)/mouse is the best dose to evaluate the protective effect of the lethal model of MRSA252 bacterial infection.
- Strain recovery: Obtain a tube of frozen MRSA252, inoculate the bacteria on Mueller Hinton (MH) (A) plates (see Table of Materials) by the "three-line method", and culture overnight at 37 °C. Perform the steps of the "three-line method" as mentioned below:
- Holding the inoculation ring in the right hand, cauterize and cool it to obtain a ring of bacterial liquid.
- Hold the agar plate in the left hand (keeping the lid on the table) above the front left area of the flame of the alcohol lamp so that the plate faces the flame, preventing the bacteria from entering the air. With the right hand, move the inoculation ring of infected bacteria back and forth on the agar surface in a dense but nonoverlapping way, covering an area of approximately 1/5-1/6 of the whole plate, which is the first zone.
NOTE: The inoculation ring and the agar should be in gentle contact at a 30°-40° angle; damaging the agar can be avoided by sliding with wrist force. - Cauterize the excess bacteria on the inoculation ring (whether to cauterize or sterilize each area depends on the amount of bacteria in the specimen). After cooling, repeat the penultimate Rows 2-3 at the end and draw a second area (about 1/4 of the plate area).
- After the second zone is finished, cauterize the ring, and draw the third zone in the same way to fill the entire plate.
- After the lines are drawn, place the plate on the lid of the dish, mark the plate, and incubate in a 37 °C incubator for 18-24 h to observe the distribution of colonies on the agar surface. Focus attention on whether a single colony is isolated, and observe colony characteristics (such as size, shape, transparency, and pigmentation).
- Bacterial activation: The next day, take two 50 mL shaker flasks and add 20 mL of MH (B) medium (see Table of Materials) to each. Pick two single colonies with a 10 µL pipette tip and add them to the shakers. Incubate the colonies overnight at 220 rpm at 37 °C.
- Secondary activation: The next day, take two 50 mL shaker flasks, each containing 20 mL of MH (B) medium. Remove 200 µL of bacterial solution from the shaker containing the first activated bacteria, add to the two new flasks containing 20 mL of MH (B) medium, and incubate at 37 °C and 220 rpm for 5 h.
- Collect the second activated bacterial solution in a 50 mL centrifuge tube, separate at 3,000 x g for 20 min, and discard the supernatant.
- Resuspend the precipitated bacteria in 40 mL of saline.
- Measure the OD600 value of the bacterial solution and adjust to 1 with saline. At this time, the bacterial quantity of MRSA252 in our experiment was 2 x 109 CFU/mL.
- Dilute the bacterial solution with an OD600 of 1 to a concentration of 1 x 109 CFU/mL.
- Randomly divide 48 Balb/c mice into four groups (n=12).
NOTE: Group I: PBS group; Group II: BNE control group; Group III: naive antigen [protein] group; Group IV: nanoemulsion adjuvant vaccine group. - Anesthetize the mice by intraperitoneal injection of 100 mg/kg of 1% pentobarbital sodium.
- Irradiate the mice with an infrared physiotherapy lamp (see Table of Materials) for approximately 1 min to cause vasodilation of the tail vein.
- Challenge the mice with injection of the diluted bacterial solution (step 5.7) into the tail vein, 100 µL per mouse.
- Place the mice under the infrared physiotherapy lamp until they can return to normal activity.
- Observe and record the survival of mice every 12 h for 10 days.
- Euthanize the mice that survived the attack by intraperitoneal injection of 100 mg/kg of 1% sodium pentobarbital.
Representative Results
The protocol for preparing the nanoemulsion adjuvant vaccine and in vitro and in vivo tests of this vaccine was evaluated. TEM, AFM, and DLS were used to determine the important characteristics of the zeta potential and particle size on the surface of this sample (Figure 1). SDS-PAGE and western blotting showed that the amount of antigen in the precipitate and supernatant did not significantly degrade after centrifugation, which indicated that the vaccine was structurally intact, specific, and immunogenic (Figure 2). The nanoemulsion adjuvant vaccine significantly increased the total IgG, IgG1, and IgG 2a antibody titers. The immunogenicity of the vaccine was significantly improved (Figure 3A–C). The serum IgG2b OD values at 450 nm of the antigen group were the highest (dilute at 1:500 PBS) (Figure 3D). The lethal MRSA252 mouse model most directly reflected that the nanoemulsion adjuvant vaccine showed a good protective effect and effectively inhibited bacterial infection with MRSA252, improving the survival rate of infected mice (Figure 4).
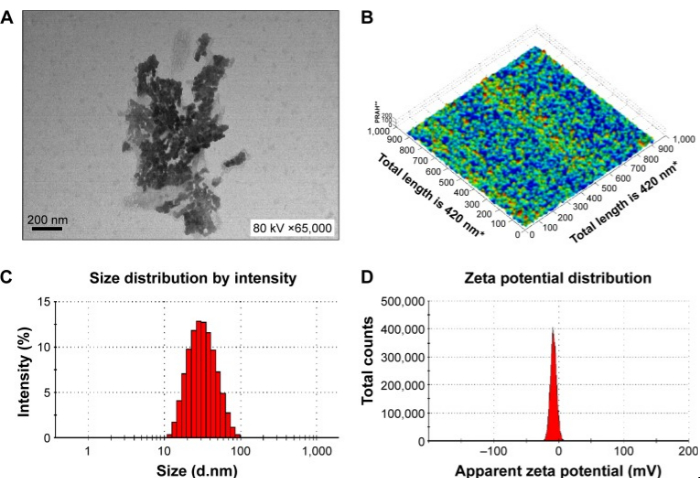
Figure 1: Physical characteristics of the novel nanoemulsion adjuvant vaccine. (A) Transmission electron micrograph. (B) Atomic force microscopy micrograph. (C) Size diameter and distribution. (D) Zeta potential and distribution. The figure is reproduced from Sun, H. W. et al. International Journal of Nanomedicine. 10, 7275-7290 (2015)11. Originally published by and used with permission from Dove Medical Press Ltd. Please click here to view a larger version of this figure.
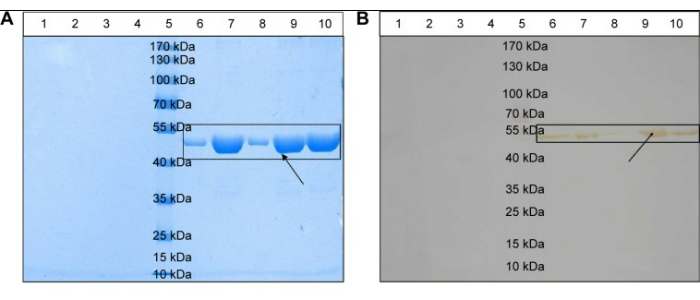
Figure 2: Physical stability of the novel nanoemulsion adjuvant vaccine. (A) Structural integrity of the antigenic proteins. (B) Structural specificity of the antigen protein. Lane 3: blank nanoemulsion-treated supernatant; Lane 4: blank nanoemulsion treatment precipitate; Lane 5: prestained marker; Lane 6: vaccine supernatant; Lane 7: vaccine precipitate; Lane 8: vaccine treatment supernatant; Lane 9: vaccine treatment precipitate; Lane 10: native protein agent; (A) and (B) mixed term. Clearly defined protein channels are marked by black squares. Black arrows indicate antigenic proteins. The figure is reproduced from Sun, H. W. et al. International Journal of Nanomedicine. 10, 7275-7290 (2015)11. Originally published by and used with permission from Dove Medical Press Ltd.Sun et al. Please click here to view a larger version of this figure.
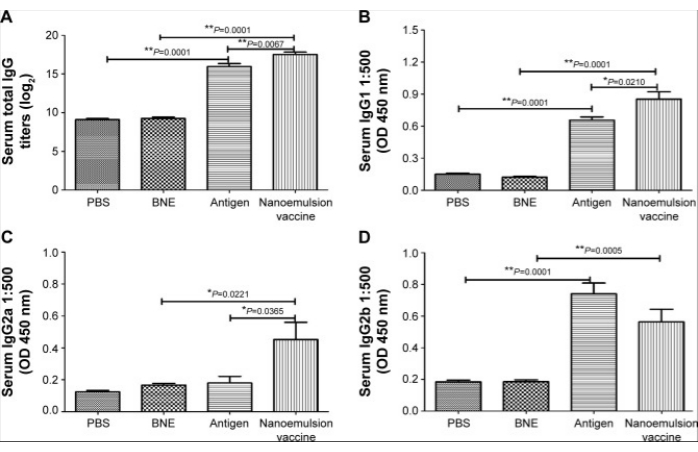
Figure 3: IgG and its subgroup antibody levels after intramuscular injection with the nanoemulsion adjuvant vaccine. (A) Log2 value of the IgG titers. (B) 450 nm optical density of serum IgG1. (C) 450 nm optical density of serum IgG2a. (D) 450 nm optical density of serum IgG2b. The figure is reproduced from Sun, H. W. et al. International Journal of Nanomedicine. 10, 7275-7290 (2015)11. Originally published by and used with permission from Dove Medical Press Ltd. Please click here to view a larger version of this figure.
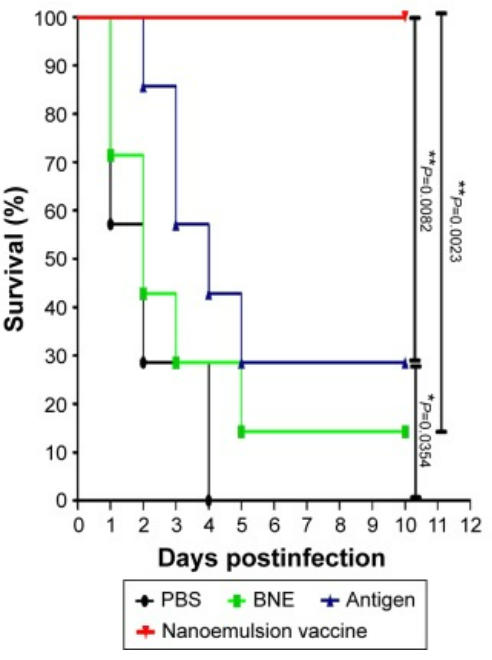
Figure 4: Protective effect in the lethal model of MRSA252 bacterial infection. The survival ratio of mice in response to systemic MRSA infection. The figure is reproduced from Sun, H. W. et al. International Journal of Nanomedicine. 10, 7275-7290 (2015)11. Originally published by and used with permission from Dove Medical Press Ltd. Please click here to view a larger version of this figure.
Table 1: ELISA spiking sequence and dilution template. Please click here to download this Table.
Discussion
IsdB, a bacterial cell wall-anchored and iron-regulated surface protein, plays an important role in the process of obtaining heme iron15. Hla, alpha toxin, is among the most effective bacterial toxins known in MRSA, and can form pores in eukaryotic cells and interfere with adhesion and epithelial cells16. In our study, a novel recombination MRSA antigen protein (HI) was constructed and expressed with gene engineering technology based on the antigen genes of IsdB and Hla. Then, a nanoemulsion vaccine was developed by the low-energy emulsification method, and the stability and effectiveness of this vaccine were evaluated. For example, some characteristics, such as long-term stability, structure, shape, absorption, and effect in vivo, are greatly affected by the size of the emulsion droplet7. The results showed that particle size was an important factor affecting the transport of nanoemulsions in vivo and their phagocytosis by antigen-presenting cells17. Regarding cytophagocytosis, nanoparticles smaller than 1 µm are more easily phagocytosed by dendritic cells (DCs) and macrophages and are the most likely to stimulate systemic immunity18.
Regarding particle transport, nanoparticles smaller than 100 nm are more likely to be transmitted through lymphatic vessels to enhance the immune effect; however, small nanoparticles infiltrate into blood vessels, while large nanoparticles slow their transport speed in lymphatic vessels19. Thus, the optimal particle size is in the range of 20-50 nm. Hence, it is critical to study the physical characteristics of nanoparticles, including shape, size, and zeta potential. TEM, with its high precision, has become an important and basic tool for understanding the properties of nanoemulsion adjuvant vaccines. Additionally, the technology has been widely applied in many fields for several years. Another nanomechanical characterization tool with 3D and nanoscale high-resolution information, AFM, was also used to evaluate the nanoemulsion vaccine13. Importantly, DLS, which simultaneously reveals both size and charge20, can provide key information, such as the aggregation state of nanoemulsion vaccine particles in solution. According to the literature, a high zeta potential value is indispensable for the physical and chemical stability of colloidal suspensions, as a large repulsive force often prevents aggregation caused by occasional collisions with adjacent nanoparticles. Therefore, in our research, we set up these schemes and evaluated their physical and chemical properties with TEM, AFM, and DLS.
Protein vaccines play an important role in the prevention and treatment of major infectious diseases21. Protein vaccines have developed rapidly in recent years, but serious challenges remain in immune efficacy and persistence. The fundamental reasons for this are poor protein stability, small distribution coefficient, short biological half-life of antigens, and high clearance rates in vivo22. In this protocol, the stability of the novel nanoemulsion vaccine was evaluated, and no antigen degradation was observed by SDS-PAGE or western blot.
A safe, effective, and marketable protein vaccine must be supplemented with an appropriate immune adjuvant to significantly enhance the immunogenicity of the protein antigen and the type of protective immune response20 to obtain the optimal vaccine immune protection. The antibody level stimulated by a protein vaccine is among the important indicators for the effective prevention and treatment of infection, and the antibody titer is an important index to evaluate whether the novel nanoemulsion adjuvant can improve the immunogenicity of the naked antigen. Novel immune adjuvants can significantly stimulate the body to enhance the humoral and cellular immune response levels, and enriching the types of immune response and immune protective efficacy of vaccines is an important development direction and the component of future research and the development of novel vaccines23. Therefore, in this protocol, antibody titers of mouse serum-specific IgG, IgG1, IgG2a, and IgG2b with the novel nanoemulsion adjuvant vaccine were detected. The lethal mouse model24 is the gold standard for evaluating vaccines, and in our protocol, a lethal MRSA252 mouse model was used to evaluate the protective effect. The results showed that the novel nanoemulsion adjuvant vaccine effectively improved the survival rate of Balb/c mice infected with MRSA.
This protocol is not sufficient in the stability evaluation of nanoemulsion vaccines. Circular dichroism is a suitable method if it can be applied to detect the spatial structure of vaccine proteins in a simple and fast method. Although the survival rate of mice tested by the attack is the gold standard for evaluating the in vivo test and effectiveness of the vaccine, it would be more convincing if the amount of bacterial colonization of the organs of mice after attack could be measured. Therefore, there are still some shortcomings in the present protocol to evaluate the immune response of the nanoemulsion adjuvant vaccine, and the above experiment needs to be supplemented in subsequent relevant experiments for evaluation.
In conclusion, it is necessary to establish a series of evaluations of physical properties, stability, specific immune response, and challenge protection. Completing these schemes will provide vital experimental fundamentals for the application and development of novel nanoemulsion adjuvant vaccines.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported by No. 2021YFC2302603 of the National Key Research and Development Program of China, No. 32070924 and 32000651 of the NSFC, and No. 2019jcyjA-msxmx0159 of the Natural Science Foundation Project Program of Chongqing.
Materials
| 5424-Small high speed centrifugeFA-45-24-11 | Eppendorf, Germany | 5424000495 | |
| 96-well plates | Corning Incorporated, USA | CLS3922 | |
| AFM Dimension FastScan | BRUKER, Germany | null | |
| Alcohol lamp | Shenzhen Yibaxun Technology Co.,China | YBS-AA-11408 | |
| Balb/c mice | Beijing HFK Bioscience Co. Ltd. | ||
| BCIP/NBT | Fuzhou Maixin Biotechnology Development Company,China | BCIP/NBT | |
| Bio-Rad 6.0 microplate reader | Bio-Rad Laboratories Incorporated Limited Co., CA, USA | null | |
| BL21 Competent Cell | Merck millipore,Germany | 70232-3CN | |
| BSA-100G | Sigma-Aldrich, USA | B2064-100G | |
| Centrifuge 5810 R | Eppendorf, Germany | 5811000398 | |
| Coomassie bright blue G-250 staining solution | MIKX,China | DB236 | |
| Decolorization solution | BOSTER,China | AR0163-2 | |
| Electro-heating standing-temperature cultivator HH-B11-420 | Shanghai Yuejin Medical Device Factory, China | null | |
| Electrophoresis apparatus | Beijing Liuyi Instrument Factory, China | DYCZ-25D | |
| Gel image | Tanon, USA | null | |
| Glutathione-Sepharose Resin GST | Mei5bio,China | affinity chromatography resin | |
| H2SO4 | Chengdu KESHI Chemical Co., LTD,China | 7664-93-9 | |
| HI recombinant protein | Third Military Medical University,China | 110-27-0 | |
| HRP -Goat Anti-Mouse IgG | Biodragon, China | BF03001 | |
| HRP- Goat anti-mouse IgG1 | Biodragon, China | BF03002R | |
| HRP- Goat anti-mouse IgG2a | Biodragon, China | BF03003R | |
| HRP- Goat anti-mouse IgG2b | Biodragon, China | BF03004R | |
| Inoculation loop | Haimen Feiyue Co.,LTD,China | YR-JZH-1UL | |
| IsdB and Hla clones | Shanghai Jereh Biotechnology Co,China | null | |
| Isopropyl nutmeg (pharmaceutic adjuvant) | SEPPIC, France | null | |
| isopropyl- β-D-1-mercaptogalactopyranoside | fdbio,China | FD3278-1 | |
| LB bouillon culture-medium | Beijing AOBOX Biotechnology Co., LTD,China | 02-136 | |
| Lnfrared physiotherapy lamp | Guangzhou Runman Medical Equipment Co.,China | 7600 | |
| Low temperature refrigerated centrifuge | Eppendorf, Germany | null | |
| Malvern NANO ZS | Malvern Instruments Ltd., UK | null | |
| MH(A) medium | Beijing AOBOX Biotechnology Co., LTD,China | 02-051 | |
| MH(B) medium | Beijing AOBOX Biotechnology Co., LTD,China | 02-052 | |
| Micro plate washing machine 405 LSRS | Bio Tek Instruments,Inc Highland Park,USA | null | |
| Mini-TBC Compact Film Transfer Instrument | BeiJingDongFangRuiLi Co.,LTD,China | 1658030 | |
| MMC packing | TOSOH(SHANGHAI)CO.,LTD | 0022818 | |
| MRSA252 | USA, ATCC | null | |
| Nanodrop ultraviolet spectrophotometer | Thermo Scientific, USA | null | |
| New FlashTM Protein any KD PAGE Protein electrophoresis gel kit | DAKEWE, China | 8012011 | |
| PBS | biosharp, China | null | |
| PCR, Amplifier | Thermal Cycler, USA | null | |
| pGEX-target gene recombinant plasmid | Shanghai Jereh Biotechnology Co,China | B3528G | |
| Phosphotungstic acid | G-CLONE, China | CS1231-25g | |
| pipette | Eppendorf, Germany | 3120000844 | |
| polyoxyethylated castor oil (pharmaceutic adjuvant) | Aladdin, China | K400327-1kg | |
| Primary antibody | Laboratory homemade:from immunized mice with positive sera | null | See Reference 11 for details |
| propylene glycol (pharmaceutic adjuvant) | Sigma-Aldrich, USA | P4347-500ML | |
| Protein Marker | Thermo Scientufuc, USA | 26616 | |
| PVDF TRANSFER MEMBRANE | Invitrogen,USA | 88518 | |
| Scanning Electron Microscope | JEOL,Japan | JSM-IT800 | |
| Sodium pentobarbital | Merck,Germany | Tc-P8411 | |
| Talos L120C TEM | Thermo Fisher, USA | null | |
| TMB color solution | TIAN GEN, China | PA107-01 | |
| Turtle kits | Xiamen Bioendo Technology Co.,LTD | ES80545 | |
| Tween-20 | Macklin, China | 9005-64-5 |
References
- Cheung, G. Y. C., Bae, J. S., Otto, M. Pathogenicity and virulence of Staphylococcus aureus. Virulence. 12 (1), 547-569 (2021).
- Lakhundi, S., Zhang, K. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clinical Microbiology Reviews. 31 (4), e00020 (2018).
- Mancuso, G., Midiri, A., Gerace, E., Biondo, C. Bacterial antibiotic resistance: the most critical pathogens. Pathogens. 10 (10), 1310 (2021).
- Goullé, J. P., Grangeot-Keros, L. Aluminum and vaccines: Current state of knowledge. Medecine et Maladies Infectieuses. 50 (1), 16-21 (2020).
- Shi, S., et al. Vaccine adjuvants: Understanding the structure and mechanism of adjuvanticity. Vaccine. 37 (24), 3167-3178 (2019).
- Geoghegan, S., O’Callaghan, K. P., Offit, P. A. Vaccine safety: myths and misinformation. Frontiers in Microbiology. 11, 372 (2020).
- Pandey, P., Gulati, N., Makhija, M., Purohit, D., Dureja, H. Nanoemulsion: a novel drug delivery approach for enhancement of bioavailability. Recent Patents on Nanotechnology. 14 (4), 276-293 (2020).
- Ko, E. J., Kang, S. M. Immunology and efficacy of MF59-adjuvanted vaccines. Human Vaccines & Immunotherapeutics. 14 (12), 3041-3045 (2018).
- Chen, B. H., Inbaraj, B. S. Nanoemulsion and nanoliposome based strategies for improving anthocyanin stability and bioavailability. Nutrients. 11 (5), 1052 (2019).
- Zuo, Q. F., et al. Evaluation of the protective immunity of a novel subunit fusion vaccine in a murine model of systemic MRSA infection. PLoS One. 8 (12), e81212 (2013).
- Sun, H. W., et al. Induction of systemic and mucosal immunity against methicillin-resistant Staphylococcus aureus infection by a novel nanoemulsion adjuvant vaccine. International Journal of Nanomedicine. 10, 7275-7290 (2015).
- National Pharmacopoeia Committee. . Chinese Pharmacopoeia. , 1088 (2020).
- Kontomaris, S. V., Stylianou, A., Malamou, A. Atomic force microscopy nanoindentation method on collagen fibrils. Materials. 15 (7), 2477 (2022).
- Zeng, H., et al. An immunodominant epitope-specific monoclonal antibody cocktail improves survival in a mouse model of Staphylococcus aureus bacteremia. The Journal of Infectious Diseases. 223 (10), 1743-1752 (2021).
- Roy, U., Kornitzer, D. Heme-iron acquisition in fungi. Current Opinion in Microbiology. 52, 77-83 (2019).
- Saeed, K., et al. Bacterial toxins in musculoskeletal infections. Journal of Orthopaedic Research. 39 (2), 240-250 (2021).
- Xu, Q., Zhou, A., Wu, H., Bi, Y. Development and in vivo evaluation of baicalin-loaded W/O nanoemulsion for lymphatic absorption. Pharmaceutical Development and Technology. 24 (9), 1155-1163 (2019).
- Singh, Y., et al. Nanoemulsion: Concepts, development and applications in drug delivery. Journal of Controlled Release. 252, 28-49 (2017).
- Kadakia, E., Shah, L., Amiji, M. M. Mathematical modeling and experimental validation of nanoemulsion-based drug transport across cellular barriers. Pharmaceutical Research. 34 (7), 1416-1427 (2017).
- Bhattacharjee, S. DLS and zeta potential-What they are and what they are not. Journal of Controlled Release. 235, 337-351 (2016).
- Francis, M. J. Recent advances in vaccine technologies. The Veterinary Clinics of North America. Small Animal Practice. 48 (2), 231-241 (2018).
- Tripathi, N. K., Shrivastava, A. Recent developments in recombinant protein-based dengue vaccines. Frontiers in Immunology. 9, 1919 (2018).
- Wilder-Smith, A. Dengue vaccine development: status and future. Bundesgesundheitsblatt Gesundheitsforschung Gesundheitsschutz. 63 (1), 40-44 (2020).
- Korneev, K. V. Mouse models of sepsis and septic shock. Molecular Biology. 53 (5), 799-814 (2019).

