A Quantitative Cell Migration Assay for Murine Enteric Neural Progenitors
Summary
We present an ex vivo cell migration assay that allows precise quantification of enteric neural crest cell migration potential in the presence of various growth factors.
Abstract
Neural crest cells (NCC) are a transient and multipotent cell population that originates from the dorsal neural tube and migrates extensively throughout the developing vertebrate embryo. In addition to providing peripheral glia and neurons, NCC generate melanocytes as well as most of the cranio-facial skeleton. NCC migration and differentiation is controlled by a combination of their axial origin along the neural tube and their exposure to regionally distinct extracellular cues. Such contribution of extracellular ligands is especially evident during the formation of the enteric nervous system (ENS), a complex interconnected network of neural ganglia that locally controls (among other things) gut muscle movement and intestinal motility. Most of the ENS is derived from a small initial pool of NCC that undertake a long journey in order to colonize – in a rostral to caudal fashion – the entire length of the prospective gut. Among several signaling pathways known to influence enteric NCC colonization, GDNF/RET signaling is recognized as the most important. Indeed, spatiotemporally controlled secretion of the RET ligand GDNF by the gut mesenchyme is chiefly responsible for the attraction and guidance of RET-expressing enteric NCC to and within the embryonic gut. Here, we describe an ex vivo cell migration assay, making use of a transgenic mouse line possessing fluorescently labeled NCC, which allows precise quantification of enteric NCC migration potential in the presence of various growth factors, including GDNF.
Introduction
Neural crest cells (NCC) are a transient cell type unique to vertebrates that forms many derivatives during embryo development. This cell population arises at the border of the neural plate, adjacent to non-neural ectoderm1. During neurulation, bending of the neural plate places NCC along the dorsal edge of the forming neural tube. NCC then undergo an epithelial-mesenchymal transition, segregating and migrating away from the neural tube. NCC colonize various embryonic structures, including the digestive tract where they form the entire enteric nervous system (ENS), an interconnected network of neural ganglia embedded in the intestinal wall. As recently reviewed2,3, many genes have been involved in the development of this intricate structure.
Most of the ENS is derived from a small pool of NCC originating from the vagal neural tube (i.e. around the prospective hindbrain/spinal cord boundary)4. These neural progenitors reach the foregut around embryonic day (e) 9.0 in mice and then migrate caudally within the gut mesenchyme until approximately e15.0 to colonize the whole embryonic intestines. A minor subset of colonic neural progenitors is also provided by sacral NCC, which invade the posterior gut in the opposite direction up to the cecum4. Both vagal and sacral NCC require multiple migration-, proliferation-, survival- and differentiation-promoting cues to ensure complete formation of the ENS. In this regard, animal models – especially genetically modified mice – have been instrumental in the identification of several essential extracellular ligands: GDNF (glial cell-derived neurotrophic factor), Endothelin-3, Neurotrophin-3, BMPs (bone morphogenic proteins), Netrin, as well as Sonic and Indian Hedgehog (Shh and Ihh)5-10. Of these, GDNF signaling through the tyrosine kinase transmembrane receptor RET (Rearranged during transfection) is recognized as the most critical pathway for the attraction and guidance of NCC to and within the embryonic gut. GDNF is secreted by the gut mesenchyme and forms a spatiotemporally controlled rosrrocaudal gradient that is directly chemoattractive to enteric NCC, which express RET11,12.
Amongst other functions, the ENS regulates movement within the digestive tract through its interaction with smooth muscle in the intestinal wall. Absence of neural ganglia in the terminal region of the bowel results in Hirschsprung's disease: tonic contraction of the affected segment leads to blockage, upstream accumulation of digested material and massive distention of the gut and abdomen. Hirschsprung's disease occurs approximately one in 5,000 live births. The rostro-caudal migration pattern of enteric NCC is believed to be the main contributing factor to the etiology of Hirschsprung's disease. The colon, furthest from the source of migrating NCC and last portion of bowel to be colonized, is most susceptible to defects in ENS formation. In accordance with its crucial role in enteric NCC migration, disruption of GDNF/RET signaling is the main known genetic cause of Hirschsprung's disease13.
To better study NCC and ENS development, we generated a transgenic mouse line – named Gata4p[5kb]-GFP14 – in which migratory NCC are labeled with Green Fluorescent Protein (GFP). We next perfected an ex vivo cell migration assay, adapted from published work by other groups11,12,15, that now allows precise quantification of enteric NCC migration potential in the presence of various growth factors, such as GDNF.
Protocol
Ethics statement
Experiments involving mice were performed following Canadian Council of Animal Care guidelines for the care and manipulation of animals used in medical research. Protocols involving the manipulation of animals were approved by the institutional ethics committee of the University of Quebec in Montreal (Comité Institutionnel de Protection des Animaux; reference number 0512-R3-650-0513).
1. Preparation of Collagen Gels
Work in a sterile fashion, under a tissue culture hood.
- Prepare complete 5x DMEM (Eagle’s modified essential medium) including standard antibiotics. Dissolve 3.37 g of DMEM powder and 0.925 g of NaHCO3 in 20 ml water. Sterilize by passing through a 0.22 μm filter. Add 2.5 ml of sterile 100x penicillin/streptomycin and 25 ml of sterile heat-inactivated fetal bovine serum. Store at 4 °C.
- On ice, mix 800 µl of Collagen I solution (3.77 mg/ml in 0.02 N acetic acid, filter-sterilized), 600 µl of complete 5x DMEM and 17 µl of 1 N NaOH. Dilute with sterile water to a final volume of 3 ml. Include relevant growth factors in the mix. We typically use GDNF at 10 ng/ml to stimulate enteric NCC migration.
- Deposit about 480 µl in each well of a single row in a 24-well plate. Avoid bubbles. The remaining rows can be used to test the effect of other growth factors on cell migration.
- Let the collagen polymerize at least 1 hr in a sterile incubator at 37 °C.
2. Dissection of Animals16
- Set up matings and check for vaginal plugs the next morning. Day e0.5 being noon on the day the vaginal plug is found, isolate the female and wait 12 days until e12.5.
- Anesthetize pregnant mice with isoflurane and euthanize by CO2 inhalation.
- Spray the mouse with 70% ethanol. Lift the abdomen skin and open the abdomen cavity with dissecting scissors.
- Remove the uterus into a glass Petri dish filled with ice-cold PBS (phosphate-buffered saline). Cut the uterus transversely between individual deciduum swellings to isolate each embryo implantation site.
- Work on each implantation site separately in another glass Petri dish filled with ice-cold PBS. Under a dissecting microscope, use fine forceps to remove the muscle layer of the uterus.
- Open the visceral yolk sac and amnion to reveal the embryo. Take care when severing the blood vessels joining the embryo to the placenta/visceral yolk sac, as they are intertwined with the developing intestines.
- Sever the embryo's head at the neck.
- Insert a closed forceps in the abdominal cavity of an embryo, just above the dark red-colored liver and let the forceps open of itself (stop applying pressure) to make a transversal opening in the abdomen cavity. Pull the opened forceps down towards the posterior end of the embryo to open the abdomen completely.
- Grab the connective tissue behind the liver and pull the guts out of the abdomen, being careful not to break the intestines (the colon is attached to the anus).
- Cut the colon to free the guts from the rest of the embryo. The cut can be made at any location along the colon. Reserve the tail portion for later.
- Tease out the connective tissue from the cecum, then the rest of the intestines. Be careful not to wound the intestines while doing so.
- Cut away the liver and stomach (on the rostral end of the small intestine), as well as the mesonephros and genital ridges if some are present.
- Isolate the small intestine. Again, be careful not to nip the intestine. From now on, the rosrrocaudal orientation of the gut tissue can be tracked using the sharp curvature present on the rostral end. Leave the small intestine in PBS at room temperature for as short a time as possible before embedding (see step 3.3).
- Finally, record the number of tail somites for each embryo to accurately determine the stage of embryonic development.
3. Sectioning of Embryonic Intestines
- Before proceeding with embryo dissections, melt 1.5 g agarose in 100 ml PBS (phosphate-buffered saline) and keep at 50 °C.
- Pour melted agarose into an embedding mold (e.g. a closed 2 ml microcentrifuge tube that has been cut lengthwise to excise about 1/4 of the tube wall). Let the agarose cool down to approximately 42-45 °C, it should be only slightly warm to the touch.
- Embed the embryonic intestine just before the agarose solidifies (this can be evaluated with forceps tips and will occur around 36-38 °C). Holding the intestine with forceps by the folded rostral end, pull it very slowly through the agarose along the length of the mold. This helps to keep the gut straight while the agarose sets. Release the tissue as soon as it begins to resist being moved. Keep track of the rostral-caudal orientation of the gut.
- Put the mold in a refrigerator 2-3 minutes, to ensure the agarose has completely set.
- Take out the agarose from the mold (by sliding it out of the opened Eppendorf tube). With a blade, take out the excess agarose at both ends, making cuts perpendicular to the intestine.
- Glue the rostral end of the intestine/agarose block down on the metal stage of a microtome with vibrating blade. Trim the excess agarose on the sides of the intestine/agarose block.
- Mount the metal stage on the vibrating microtome chamber. If necessary, adjust the angle of the stage so the intestine is as vertical as possible (hence perpendicular to the blade). Cover the specimen with ice-cold PBS. Bring the microtome blade down to a few millimeters under the buffer surface.
- Make 200 μm vibratome transverse cuts of the caudal-most small intestine, ensuring that each agarose slice contains a full intestinal section.
4. Culture of Intestinal Explants
- Gently deposit the freshly cut intestine/agarose slices flat onto the collagen gels with forceps, placing one slice towards the middle of each well.
- Incubate 3 days at 37 °C, in a humid 5% CO2 atmosphere, to allow migration of NCC out of the explant.
- Take the intestine/agarose slices off the collagen gel very gently with forceps. Take care not to damage the gel below.
- Avoid touching the collagen gel directly during the subsequent incubation and wash steps in order to ensure that the cell migration pattern is not disturbed. Fix with 500 µl of 4% PFA (paraformaldehyde) (in PBS) per well 1 hr at room temperature.
- Replace the fixative with 500 µl of DAPI (4′,6-diamidino-2-phenylindole) solution (5 µg/ml in PBS) per well and incubate 10 min at room temperature.
- Wash each well 3x with 500 µl of PBS for 5 min.
- Photograph the fluorescent cells (GFP and DAPI channels) embedded in the collagen gel within each well.
5. Image Analysis
We made extensive use of ImageJ17 to process and quantify the images generated after explant culture.
- Start by imaging a micrometer slide at the same magnification as the fluorescent cell photographs. Measure the length of one micron in pixels (using the Straight Line tool).
- Set the scale (Analyze/Set Scale; input number of pixels/micron).
- For each GFP fluorescence photograph, change the image format to 8-bit grayscale (Image/Type/8-bit).
- Adjust intensity of the signal (Image/Adjust/Brightness/Contrast).
- Subtract the background noise if necessary (Process/Subtract Background; adjust the rolling ball radius).
- Set a threshold to highlight cells and cell clumps (Image/Adjust/Threshold), then apply a watershed to divide the clumps (Process/Binary/Watershed).
- Analyze particles to specify regions of interest (ROI) and generate cell number statistics (Analyze/Analyze Particles; set size minimum to exclude remaining pixels).
- Set measurement options to include Feret's diameter (Analyze/Set Measurements).
- Group ROIs and measure as a whole to determine the Feret's diameter, an indication of cell spread (Analyze/Tools/ROI Manager/More/OR, then ROI Manager/Add, and ROI Manager/Measure).
Representative Results
The following results are representative of what can be obtained with the technique described here (Figure 1). The use of growth factors (i.e. GDNF) stimulates migration of GFP-expressing enteric NCC out of the intestinal explant and into the collagen gel (Figure 2). Though some cells come out of the explant in the absence of growth factors, these are mostly not GFP-labeled and represent passive entry. It is necessary to remove the intestinal slice from the collagen gel in order to record the results, as this tissue is still heavily populated by fluorescent cells and would otherwise hide the cells lying in the collagen underneath. Quantification of these results shows that many more cells are found within the collagen gel when GDNF is present, and that active migration takes place (Figure 3). Indeed, passive cells are found immediately under the explants, within the diameter of an intestinal slice, whereas cells that actively invade the collagen gel move away from their point of origin and spread further.
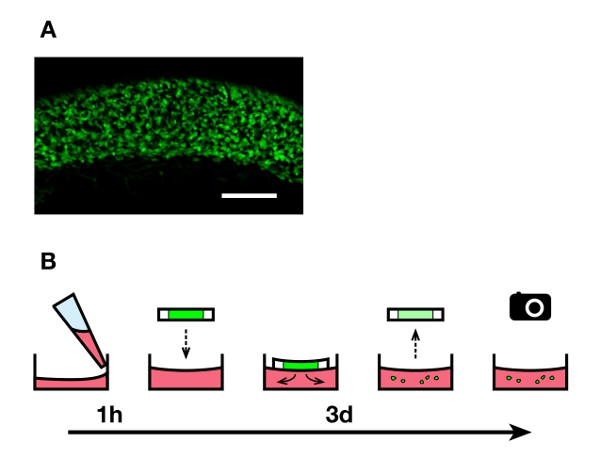
Figure 1. Overview of the explant culture technique. A) Uniform population of fluorescent enteric NCC within the caudal small intestine region used to make 200 μ-thick explants. Scale bar: 200 μm. B) The culture medium containing collagen is deposited in 24-well plates and left to harden for 1 hr. Vibratome slices of agarose-embedded embryonic intestines are deposited on the gels (one slice per well), and fluorescent enteric NCC are allowed to migrate out of the explants for 3 days. The slices are then taken off before imaging the cells that invaded the collagen gels.
Click here to view larger image.
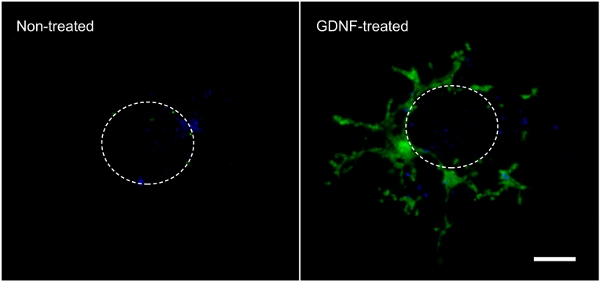
Figure 2. Cell migration out of the intestinal explant and within the collagen gel. Cells that migrated out of an intestinal explant during the 3-day incubation in the absence or presence of 10 ng/ml GDNF were fixed, stained with DAPI (blue) and photographed to show GFP-labeled enteric NCC (green). 70X magnification. Scale bar: 100 μm. The dotted line represents the approximate size and location of the explant before it was taken off the gel. Click here to view larger figure.
Click here to view larger image.
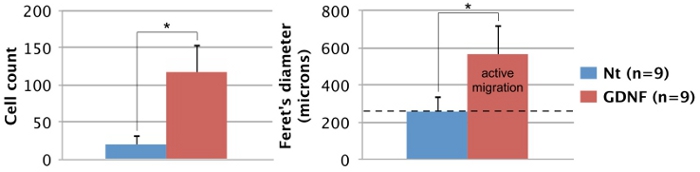
Figure 3. Quantification of enteric NCC migration potential. The number and spread (Feret's diameter) of GFP-expressing cells migrating out of intestinal explants after 3 days was quantified using ImageJ software17. In both cases, there is a significant difference between untreated and GDNF-treated conditions according to a Student’s t test (p<0.001; *). The average diameter of intestinal slices (dotted black line: 260 μm) was included to distinguish between passive entry and active migration. Nt: non-treated, n: number of explants processed.
Click here to view larger image.
Discussion
We show how our ex vivo explant culture technique can be used to precisely quantify enteric NCC migration potential in the presence of GDNF. Such precise quantification is greatly facilitated by using 200 μm-thick vibratome gut sections instead of large pieces of approximate size, as previously described11,12,15. Indeed, this allows us to work with a reasonable number of cells in a highly reproducible setting. Of note, the uniform distribution of fluorescent enteric NCC within the caudal-most region of the small intestine from which the explant slices are cut also allows analysis of multiple sections from a single gut (Figure 1A). Moreover, given that both enteric NCC and axons can exit the tissue in such assays11, withdrawal of the explants at the end of culture period lets us focus exclusively on migratory NCC.
Most critical steps were outlined in the protocol text, however as the welfare of the intestinal explant is paramount to obtaining healthy migrating NCC, particular care should be taken. Avoid subjecting embryonic tissues to sudden temperature changes, particularly when the intestine is embedded in agarose (step 3.3). Make sure the intestine is at room temperature and the agarose as cool as possible (yet still melted) to avoid "cooking" the tissue. The intestinal explant should thrive on the culture medium-filled collagen gel, often increasing in size and spilling out of the agarose slice. If the explant appears unhealthy or worse, tends to perish during incubation, try replacing the PBS with culture media at room temperature (e.g. HEPES-buffered M2 or DMEM supplemented with 10% FBS) to help sustain it during dissection.
A major limitation to our approach is that it relies on the availability of a mouse line conferring a fluorescent label to migrating NCC. In the absence of such a resource, an antibody against migrating NCC (e.g. anti-Ret or anti-Sox10) can be used to label cells that invaded the collagen gel. Moreover, given that the gut micro-environment is far more complex than a simple collagen gel, results obtained with this in vitro assay might not entirely reflect the behavior of enteric NCC in vivo. Additional experiments involving live-cell imaging are recommended to assess this behavior. It is also noteworthy that in addition to its role as a chemoattractant, GDNF is known to promote proliferation of migrating enteric NCC4. Our measure of enteric NCC migration potential in the presence of GDNF is thus probably a mix of true cell migration and cell proliferation, akin to the in vivo mechanisms leading to NCC colonization of the intestines. If a clear distinction between these two processes is desired, the addition of a cell cycle blocker (e.g. AZD 543818) in the culture media can restrict the analysis to cell migration.
This technique can be expanded to test various other extracellular ligands as well as inhibitors of specific signaling pathways, and any combination therein. Other tissues can also potentially be dissected and sectioned, allowing the study of NCC migration in many embryonic structures. Combined with novel and/or uncharacterized mutant mouse strains with possible defects in NCC development, our technique can be applied to quickly screen for deficiencies in migration behavior in response to specific signaling events.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We thank Denis Flipo for image processing and analysis advice, and David W. Silversides in whose laboratory the Gata4p[5kb]-GFP mouse line was generated. Research in the Pilon laboratory is funded by CIHR, NSERC, FRQS and FRQNT.
Materials
| DMEM powder | Wisent | 219-010-XK | |
| NaHCO3 | Bioshop | SOB999 | Biotechnology grade |
| Steriflip vacuum filtration system (0.22 micron) | EMD Millipore | SCGP00525 | |
| Penicilin-Streptomycin solution, 100x | Wisent | 450-201-EL | |
| Fetal bovine serum | Wisent | 095-150 | High quality grade |
| Collagen I | BD biosciences | 354236 | |
| NaOH | Bioshop | SHY700 | Diluted from 10 N stock then sterile-filtered |
| GDNF | Cedarlane | CLCYT305 | |
| Falcon 24-well Plate | BD biosciences | 353047 | |
| Dissecting scissors | Fisher Scientific | 089515 | |
| Glass Petri dish | VWR | 89000-306 | |
| PBS | Sigma | P5493 | Cell culture grade |
| Dissecting microscope | Leica | M125 | |
| Dumont #5 Forceps | Fine Science Tools | 11251-20 | |
| Agarose | Bioshop | AGA001 | Biotechnology grade |
| Surgical blade | Feather | 21 | |
| All Purpose Instant Krazy Glue Pen | Krazy Glue | KG824 | |
| HM 650V Vibrating-Blade Microtome | Thermo Scientific | 920110 | |
| Paraformaldehyde | Sigma-Aldrich | P6148 | |
| DAPI | Sigma-Aldrich | D9564 |
References
- Bronner, M. E., Le Douarin, N. M. Development and evolution of the neural crest: An overview. Dev. Biol. 366 (1), 2-9 (2012).
- Bergeron, K. F., Silversides, D. W., Pilon, N. The developmental genetics of Hirschsprung’s disease. Clin. Genet. 83 (1), 15-22 (2013).
- Obermayr, F., Hotta, R., Enomoto, H., Young, H. M. Development and developmental disorders of the enteric nervous system. Nat. Rev. Gastroenterol. Hepatol. 10 (1), 43-57 (2012).
- Sasselli, V., Pachnis, V., Bursn, A. J. The enteric nervous system. Dev. Biol. 366 (1), 64-73 (2012).
- Sanchez, M. P., Silos-Santiago, I., et al. Renal agenesis and the absence of enteric neurons in mice lacking GDNF. Nature. 382 (6586), 70-73 (1996).
- Baynash, A. G., Hosoda, K., et al. Interaction of endothelin-3 with endothelin-B receptor is essential for development of epidermal melanocytes and enteric neurons. Cell. 79 (7), 1277-1285 (1994).
- Chalazonitis, A., Pham, T. D., et al. Neurotrophin-3 is required for the survival-differentiation of subsets of developing enteric neurons. J. Neurosci. 21 (15), 5620-5636 (2001).
- Goldstein, A. M., Brewer, K. C., Doyle, A. M., Nagy, N., Roberts, D. J. BMP signaling is necessary for neural crest cell migration and ganglion formation in the enteric nervous system. Mech. Dev. 122 (6), 821-833 (2005).
- Jiang, Y., Liu, M. T., Gershon, M. D. Netrins and DCC in the guidance of migrating neural crest-derived cells in the developing bowel and pancreas. Dev. Biol. 258 (2), 364-384 (2003).
- Ramalho-Santos, M., Melton, D. A., McMahon, A. P. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 127 (12), 2763-2772 (2000).
- Natarajan, D., Marcos-Gutierrez, C., Pachnis, V., de Graaf, E. Requirement of signaling by receptor tyrosine kinase RET for the directed migration of enteric nervous system progenitor cells during mammalian embryogenesis. Development. 129 (22), 5151-5160 (2002).
- Young, H. M., Hearn, C. J., et al. GDNF Is a chemoattractant for enteric neural cells. Dev. Biol. 229 (2), 503-516 (2001).
- Amiel, J., Sproat-Emison, E., et al. Hirschsprung disease, associated syndromes and genetics: a review. J. Med. Genet. 45 (1), 1-14 (2008).
- Pilon, N., Raiwet, D., Viger, R. S., Silversides, D. W. Novel pre- and post-gastrulation expression of Gata4 within cells of the inner cell mass and migratory neural crest cells. Dev. Dyn. 237 (4), 1133-1143 (2008).
- Nagy, N., Goldstein, A. M. Endothelin-3 regulates neural crest cell proliferation and differentiation in the hindgut enteric nervous system. Dev. Biol. 293 (1), 203-217 (2006).
- Nagy, A., Gertsenstein, M., Vintersen, K., Behringer, R. . Manipulating the mouse embryo: a laboratory manual. , 209-250 (2003).
- Schneider, C. A., Rasband, W. S., Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9, 671-675 (2012).
- Byth, K. F., Thomas, A., et al. AZD5438, a potent oral inhibitor of cyclin-dependent kinases 1, 2, and 9, leads to pharmacodynamic changes and potent antitumor effects in human tumor xenografts. Mol. Cancer Ther. 8 (7), 1856-1866 (2009).

