Motility of Single Molecules and Clusters of Bi-Directional Kinesin-5 Cin8 Purified from S. cerevisiae Cells
Summary
The bi-directional mitotic kinesin-5 Cin8 accumulates in clusters that split and merge during their motility. Accumulation in clusters also changes the velocity and directionality of Cin8. Here, a protocol for motility assays with purified Cin8-GFP and analysis of motile properties of single molecules and clusters of Cin8 is described.
Abstract
The mitotic bipolar kinesin-5 motors perform essential functions in spindle dynamics. These motors exhibit a homo-tetrameric structure with two pairs of catalytic motor domains, located at opposite ends of the active complex. This unique architecture enables kinesin-5 motors to crosslink and slide apart antiparallel spindle microtubules (MTs), thus providing the outwardly-directed force that separates the spindle poles apart. Previously, kinesin-5 motors were believed to be exclusively plus-end directed. However, recent studies revealed that several fungal kinesin-5 motors are minus-end directed at the single-molecule level and can switch directionality under various experimental conditions. The Saccharomyces cerevisiae kinesin-5 Cin8 is an example of such bi-directional motor protein: in high ionic strength conditions single molecules of Cin8 move in the minus-end direction of the MTs. It was also shown that Cin8 forms motile clusters, predominantly at the minus-end of the MTs, and such clustering allows Cin8 to switch directionality and undergo slow, plus-end directed motility. This article provides a detailed protocol for all steps of working with GFP-tagged kinesin-5 Cin8, from protein overexpression in S. cerevisiae cells and its purification to in vitro single-molecule motility assay. A newly developed method described here helps to differentiate between single molecules and clusters of Cin8, based on their fluorescence intensity. This method enables separate analysis of motility of single molecules and clusters of Cin8, thus providing the characterization of the dependence of Cin8 motility on its cluster size.
Introduction
A large number of motility events within eukaryotic cells are mediated by the function of molecular motor proteins. These motors move along the cytoskeletal filaments, actin filaments, and microtubules (MTs), and convert the chemical energy of ATP hydrolysis into kinetic and mechanical forces required to drive biological motility within cells. The MT-based S. cerevisiae Cin8 is a bipolar, homotetrameric kinesin-5 motor protein that crosslinks and slides spindle MTs apart1. Cin8 performs essential functions during mitosis, in spindle assembly2,3,4 and spindle elongation during anaphase5,6,7. Previously, it had been demonstrated that Cin8 is a bi-directional motor, which switches directionality under different experimental conditions. For instance, under high ionic strength conditions, single Cin8 motors move toward the minus-end of the MTs, while in clusters, in multi-motor MT gliding assays, and between antiparallel MTs, Cin8 motors move mainly toward the plus-ends of the MTs8,9,10,11,12. These findings were highly unexpected because of several reasons. First, Cin8 carries its catalytic motor domain at the amino-terminus and such motors were previously believed to be exclusively plus-end directed, whereas Cin8 was shown to be minus-end directed at the single-molecule level. Second, kinesin motors were believed to be unidirectional, either minus-end or plus-end directed, whereas Cin8 was shown to be bi-directional, depending on the experimental conditions. Finally, because of the MT orientation at the mitotic spindle, the classical role of kinesin-5 motors in the separation of spindle poles during spindle assembly and anaphase B could only be explained by their plus-end directed motility on the MTs they crosslink1,13. Following the first reports on the bi-directionality of Cin8, a few other kinesin motors were demonstrated to be bi-directional14,15,16, indicating that the bi-directional motility of kinesin motors may be more common than earlier believed.
It has been previously reported that in cells, Cin8 also moves in a bi-directional manner8, supporting the notion that the bi-directional motility of some kinesin-5 motors is important for their intracellular functions. In addition, since the three kinesin-5 motors that were reported to be bi-directional are from fungal cells, a possible role for the bi-directionality of kinesin-5 motors has been recently proposed in such cells10. According to this model, in closed mitosis of fungal cells, where the nuclear envelope doesn't break down during mitosis, kinesin-5 motors provide the initial force that separates the spindle poles apart prior to spindle assembly. To perform this task, prior to spindle pole separation, kinesin-5 motors localize near the spindle poles, by their minus-end directed motility on single nuclear MTs. Once at this position, kinesin-5 motors cluster, switch directionality, capture, and cross-link MTs from neighboring spindle poles. Subsequently, kinesin-5 motors provide the initial separation of the poles by plus-end directed motility on the MTs they crosslink. By this model, both minus-end directed motility on single MTs and plus-end directed motility on cross-linked MTs during antiparallel sliding are required for fungal kinesin-5 motors to perform their roles in spindle assembly1,13.
The overall goal of the described method is to obtain high-purity fungal GFP-tagged kinesin-5 Cin8 and to perform single-molecule motility assays (Figure 1) while separately analyzing the motility of single molecules and clusters of Cin8. The separation between single molecules and clusters is important since one of the factors that had been demonstrated to affect the directionality of Cin8 is its accumulation in clusters on the MTs10,12. Alternative motility assays, such as the MT surface gliding and MT sliding assays do not provide information regarding the activity of single motor proteins17,18. The robust single-molecule motility assay and analysis methods described here have been successfully applied to characterize different aspects of kinesin-5 motors, Cin8 and Kip110,11,12,14,19,20.
Here, a detailed protocol is presented for Cin8 overexpression and purification, polymerization of MTs, and the single-molecule motility assay. Furthermore, the analyses to differentiate between single molecules and clusters of Cin8, and to determine single motor and cluster velocities by mean displacement (MD) and mean square displacement (MSD) analysis are also described. This protocol aims to help researchers to visualize all the steps of the procedures and assist with troubleshooting this type of assays.
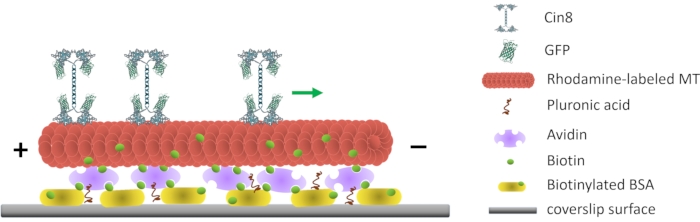
Figure 1: Schematic representation of the single-molecule motility assay. Biotinylated fluorescent MTs are attached to the glass surface, coated with Avidin that interacts with the surface-attached biotinylated-BSA. The green arrow represents the movement direction of single Cin8 molecules under high ionic strength conditions. +/- represent the polarity of the MT. Please click here to view a larger version of this figure.
Protocol
1. Preparation of buffers and reagents
- Buffers
- -Leu aa dropout mix: Mix 2 g each of Adenine, Uracil, Tryptophan, Histidine, Lysine, and Methionine and store at room temperature.
- Yeast selective medium with raffinose (1 L): Mix 6.7 g of yeast nitrogen base (with ammonium sulfate), 2 g of -Leu aa dropout mix, and 20 g of raffinose in double-distilled water by stirring (without heating) until fully dissolved. Using a 0.22 µm filter, filter the solution into a sterile bottle.
- Lysis buffer: Prepare 25 mL of solution in triple distilled water (TDW) consisting of 50 mM Tris, 30 mM Pipes, 500 mM KCl, 10% glycerol, 1.5 mM β-mercaptoethanol, 1 mM MgCl2, 0.1 mM ATP, and 0.1% Triton X-100. Adjust pH to 8 using 6 M HCl.
- Elution buffer: Prepare 10 mL of solution in TDW consisting of 50 mM Tris, 30 mM Pipes, 500 mM KCl, 350 mM imidazole, 10% glycerol, 1.5 mM β-mercaptoethanol, 1 mM MgCl2, 0.1 mM ATP, and 0.1% Triton X-100. Adjust pH to 7.2 using 6 M HCl.
- P12 Buffer: Prepare 10 mL of a solution in TDW consisting of 12 mM Pipes, 1 mM EGTA, and 2 mM MgCl2. Adjust pH to 6.9 using 10 M NaOH.
- BRB80 buffer: Prepare 50 mL of a solution consisting of 80 mM Pipes, 1 mM EGTA, and 2 mM MgCl2 in ultrapure water. Adjust pH to 6.9 using 10 M NaOH.
- General Tubulin Buffer (GTB): Prepare 50 mL of a solution consisting of 80 mM Pipes, 0.5 mM EGTA, and 2 mM MgCl2 in ultrapure water. Adjust pH to 6.9 using 10 M NaOH.
- Tris-Pipes solution: Prepare 40 mL of 1M Tris-0.6 M Pipes solution by mixing 6.055 g of Tris and 9.07 g of Pipes in TDW and adjust pH to 7.2 using 6 M HCl. Bring the final volume to 50 mL with TDW.
NOTE: P12, BRB80, and Tris-Pipes buffers are used for the preparation of stock solutions for motility assay. These buffers can be prepared in large quantities, aliquoted in 1.5 mL tubes, snap-frozen, and stored at -20 °C.
- Stock solutions for motility assay
- Tubulin (10 mg mL-1): Dissolve 1 mg of lyophilized tubulin in 100 µL of cold (4 °C) general tubulin buffer (GTB). Snap-freeze 1 µL aliquots and store them at -80 °C.
- Biotinylated tubulin (1 mg mL-1): Dissolve 20 µg of lyophilized tubulin in 20 µL of cold GTB. Snap-freeze 1 µL aliquots and store them at -80 °C.
- Rhodamine labeled tubulin (1 mg mL-1): Dissolve 20 µg of lyophilized tubulin in 20 µL of cold GTB. Snap-freeze 0.5 µL aliquots and store them at -80 °C.
- GMPCPP (10 µM): GMPCPP is obtained from the supplier as a 100 µL aqueous solution and stored at -80 °C. Thaw the vial with GMPCPP on ice. Prepare 1 µL aliquots, snap-freeze and store them at -80 °C.
- ATP: Prepare 500 µL solution of 100 mM ATP in 0.5 M Tris buffer (pH 8). Snap-freeze 2 µL aliquots and store them at -20 °C.
- MgCl2: Prepare 1 mL solution of 200 mM MgCl2 in P12 buffer. Store 5 µL aliquots at -20 °C.
- Casein: Prepare a 1 mL solution of 5 mg mL-1 casein in BRB 80 buffer. Snap-freeze 10 µL aliquots and store them at -20 °C.
- D-Glucose: Prepare a 1 mL solution of 1 M D-glucose in P12 buffer. Store 10 µL aliquots at -20 °C.
- Glucose oxidase: Prepare a 1 mL solution of 10 mg mL-1 glucose oxidase in P12 buffer. Snap-freeze 2 µL aliquots and store them at -20 °C.
- Catalase: Prepare a 1 mL solution of 0.8 mg mL-1 catalase in P12 buffer. Snap-freeze 2 µL aliquots and store at -20 °C.
- Dithiothreitol (DTT): Prepare a 1 mL solution of 1 M DTT in P12 buffer in a fume hood. Snap-freeze 10 µL aliquots and store them at -20 °C.
- Creatine phosphate: Prepare a 1 mL solution of 1 M creatine phosphate in P12 buffer. Snap-freeze 2 µL aliquots and store them at -20 °C.
- Creatine phosphokinase: Prepare a 1 mL solution of 5 mg mL-1 creatine phosphokinase in 0.25 M glycylglycine, pH 7.4. Snap-freeze 2 µL aliquots and store them at -20 °C.
- EGTA: Prepare a 100 mM EGTA solution in ultrapure water and store it at room temperature.
- KCl: Prepare a 1 M KCl solution in ultrapure water and store it at room temperature.
- Motility buffer and reaction mix
- Motility buffer (MB) with 145 mM KCl, 2x stock: Prepare 1 mL of 2x stock of the motility buffer by mixing 100 µL of pre-made Tris-Pipes solution, 20 µL of 100 mM EGTA, 290 µL of KCl, and 590 µL of TDW. Keep the buffer on ice.
- Motility reaction mix: Prepare motility reaction mix according to Table 1 and store it on ice.
| Volume | Stock | Reagent name |
| 50 µL | 2X | MB (from step 1.3.1) |
| 40 µL | – | TDW |
| 1 µL | 100 mM | ATP |
| 1 µL | 200 mM | MgCl2 |
| 2 µL | 5 mg/mL | Casein |
| 1 µL | 1 M | Glucose |
| 1 µL | 1 M | DTT |
| 1 µL | 10 mg/mL | Glucose oxidase |
| 1 µL | 8 mg/mL | Catalase |
| 1 µL | 1 M | Phosphocreatine |
| 1 µL | 5 mg/mL | Creatine phosphokinase |
| 100 µL | Total |
Table 1.
2. Cin8 overexpression and purification from S. cerevisiae cells
- Grow S. cerevisiae cells containing the plasmid for overexpression of Cin8-GFP-6His to the exponential growth phase (OD600 = 0.6-0.8) in 1 L of yeast selective medium supplemented with 2% raffinose (see step 1.1.2) at 28 °C12.
- Induce Cin8-GFP-6His overexpression by addition of 2% galactose. Monitor the yeast culture growth by measuring absorbance at 600 nm.
- Five hours after galactose addition, harvest the cells by centrifugation at 4,000 x g for 15 min at 4 °C, suspend the cells in the lysis buffer and freeze in liquid N2.
NOTE: Frozen cells can be stored at -80 °C for further use or immediately ground in liquid N2. - Grind the frozen cells in liquid N2 using chilled mortar and pestle. Add liquid N2 during the grinding to keep the extracts frozen. It typically requires 4-5 times of adding liquid N2.
- Monitor cell lysis by observation under phase-contrast or DIC microscope.
- Thaw the ground cells and centrifuge at 21,000 x g for 30 min at 4 °C. Load the supernatant onto a gravity flow column filled with 2 mL of Ni-NTA and pre-equilibrated with lysis buffer. Let the supernatant flow out through the column.
- Wash the column with five column volumes of lysis buffer, and then with five column volumes of lysis buffer supplemented with 25 mM imidazole.
- Elute Cin8-GFP-6His with elution buffer (see step 1.1.4).
- Analyze the eluted samples by SDS-PAGE fractionation, followed by Coomassie blue staining and western blot analysis probed with α-GFP antibody19.
- Pool the fractions containing Cin8-GFP-6His (steps 2.8 and 2.9). Furthermore, purify them by size-exclusion chromatography (SEC) at a flow rate 0.5 mL min-1 and column pressure limit of 1.5 MPa, with simultaneous monitoring of the absorbance at 280 nm and the GFP florescence emission with excitation at 488 nm (Figure 2A).
- Collect the fractions corresponding to the Cin8-GFP tetramer and analyze them by SDS-PAGE and western blotting (see step 2.9) (Figure 2B).
- Estimate the protein concentration using spectrophotometry or biochemical assays such as Bradford assay, BCA assay etc.
- Aliquot the selected fractions, snap-freeze in liquid N2, and store until use at -80 °C. These purified protein samples can be used for 6 months.
NOTE: Cin8-GFP is overexpressed and purified from a protease-deficient S. cerevisiae strain containing a 2 µm plasmid for Cin8-GFP-6His overexpression from the galactose inducible promoter, LGY 4093: MATα, leu2-3,112, reg-1-501, ura3-52, pep4-3, prb1-1122, gal1, pOS7 (2µ, LEU2, PGAL1–CIN8-GFP-6HIS). The yeast strain and plasmid are available upon request.
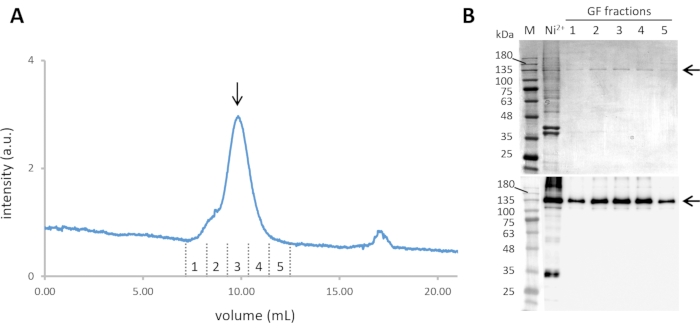
Figure 2: Purification of Cin8-GFP. (A) The size exclusion chromatogram of Ni-NTA purified Cin8-GFP, with continuous GFP fluorescence detection through 488 nm excitation and emission at ~510 nm. The Cin8-GFP tetramer elutes at ~10 mL from the SEC column (marked with an arrow). (B) Coomassie-stained SDS-PAGE gel (top) and α-GFP western blot (bottom) of Cin8-GFP fractions eluted from SEC. Samples in the lanes are as follows: M – Molecular weight marker, Ni2+– Ni-NTA purified Cin8-GFP sample that is loaded into the SEC column, GF fractions: fraction corresponding to Cin8-GFP SEC elution as marked in panel A. The arrow on the right marks the size of the Cin8-GFP monomer (expected on the SDS-PAGE). Please click here to view a larger version of this figure.
3. Single-molecule motility assay with the purified Cin8
- Polymerization of biotin and rhodamine labeled MTs, stabilized with GMPCPP.
- Start MT polymerization by mixing the following components in a 1.5 mL tube: 1 µL of 10 mg/mL tubulin protein, 1 µL of 1 mg/mL biotinylated tubulin, 0.5 µL of 1 mg/mL rhodamine-labeled tubulin, 1 µL of 10 mM GMPCPP, and 6.5 µL of general tubulin buffer (GTB). Incubate the mixture for 1 h at 37 °C.
- Following MT polymerization, add 80 µL of warm (37 °C) GTB, mix carefully and centrifuge at 16,500 x g for 20 min.
- Discard the supernatant and re-suspend the pellet carefully by pipetting up and down with 50 µL of warm GTB. Store the suspension at 28 °C.
- Examine the MTs with a fluorescence microscope using the 647 nm rhodamine channel (Figure 3A).
NOTE: To obtain biotinylated fluorescently labeled MTs, polymerization reaction contains unlabeled tubulin, as well as biotinylated and fluorescently-labeled tubulin. In this protocol, rhodamine-labeled tubulin is used but other fluorescent conjugates can be utilized as well.
- Flow Chamber assembly
- Assemble a flow chamber by placing four strips of double-sided tape (~4 cm x ~3 mm) on an advanced adhesive glass slide (parallel to the longer edge and ~3-4 mm apart) to create three 'lanes' between the tape strips. Remove the protective paper from tape strips and place a silanized coverslip10 on the tape strips to create three flow chambers of ~10 µL in volume.
- MT immobilization to the avidin-coated surface (Figure 1)
- Coat the silanized coverslip by perfusing with 15 µL of 1 mg/mL biotinylated-bovine serum albumin (b-BSA, dissolved in GTB) into the flow chamber using a micropipette. After 5 min, wash the chamber with 80 µL of GTB.
- Subsequently, as in step 3.3.1, insert into the flow chamber 15 µL of 1 mg/mL Avidin (dissolved in GTB) that binds to the b-BSA. After 5 min, wash the chamber with 80 µL of GTB.
- Passivate the silanized coverslip surface using 20 µL of 1% poloxamer. After 3 min, wash with 80 µL of GTB.
- Attach biotinylated MTs (step 3.1) to the b-BSA-avidin coated coverslip by inserting 20 µL of MTs typically diluted to 1:20 in GTB. Incubate the slides in an inverted position, i.e., with the coverslip facing downwards in a dark humidity chamber (e.g., a Petri dish containing wet tissue paper) for 5 min at room temperature. Then, wash with 200 µL of GTB.
- Apply 30 µL of motility reaction mix (see step 1.3.2) into the flow chamber.
- Dilute the Cin8-GFP motors (step 2.13) in 20 µL of motility reaction mix (see Table 1) (typically to a final concentration of 5-10 µM). Apply them to the flow chamber and immediately image the motors' movement along the MTs.
- Motor motility imaging
NOTE: MT binding and motors' motility were monitored using an epifluorescence inverted microscope equipped with a mercury arc lamp, a 100x/1.4 numerical aperture objective, and two fluorescence bandpass filter sets, one with a wavelength of 647 nm (for rhodamine) and another with a wavelength of 488 nm (for GFP).- Place a drop of immersion oil on the microscope objective. Place the flow chamber on the fluorescent microscope stage with the coverslip down facing the objective.
- Turn on the rhodamine channel to focus on the MTs attached to the coverslip surface and acquire the image with 20 ms exposure using the Micro-Manager ImageJ-Fiji software21.
- Turn on the GFP channel and acquire 90 time-lapse images with 1 s interval and 800 ms exposure, for analyzing Cin8-GFP motility.
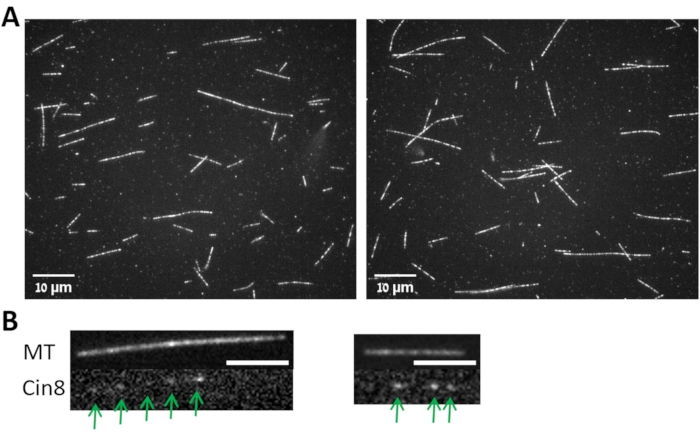
Figure 3: MTs and MT bound Cin8-GFP. (A) Images from two fields (left and right) for MTs polymerized following the protocol described in step 3.1 and imaged with 100x objective as described in step 3.4. (B) Images from two fields (left and right) for the Cin8-GFP (lower panels, marked with arrows) attached to the MT shown in the upper panels. Scale bar: 4 µm. Please click here to view a larger version of this figure.
4. Motility analysis
NOTE: Perform all the image analysis and generate kymographs using ImageJ-Fiji Software.
- Kymograph generation
- Open the time lapse movie and the corresponding MT field image. Synchronize these two windows by choosing the following option: Analyze > Tools > Synchronize Windows.
- Highlight one MT using the Segmented Line option and use the Analyze > Multi Kymograph tab to obtain a kymograph.
- Determination of cluster size of Cin8-GFP (i.e., the number of Cin8 molecules in a cluster)
- Perform the background subtraction and the correction for uneven illumination by using the Process > Subtract Background option. Set the Rolling Ball Radius at 100 pixels and check the Sliding Paraboloid option.
- Follow the mean fluorescence intensity of a specific non-motile Cin8-GFP motor (Figure 3B) as a function of time within a circle of 4 pixels radius using the TrackMate plugin of the ImageJ-Fiji software by choosing the following option: Plugins > Tracking > TrackMate > LoG Detector > Simple Lap Tracker.
- Repeat step 4.2.2 for different Cin8-GFP motors. Plot the fluorescence intensity of the different Cin8-GFP motors as a function of time.
NOTE: An experimental strategy to measure the cluster size-i.e., the number of Cin8 molecules in a cluster-establishes a basis for the analysis of Cin8 clustering-related motility. Photobleaching of GFP attached to Cin8 is employed to determine the contribution of single GFP molecules to the total intensity of Cin8 clusters. For example, the fluorescence intensities decrease in steps of ~50 arbitrary units (a.u.), with every single step probably representing the photobleaching of one GFP molecule (Figure 4A). Since Cin8 is a homo-tetrameric motor protein, it contains four GFP molecules. Thus, all Cin8 motors having an intensity ≤ 200 a.u. are likely to be single tetrameric Cin8 molecules. Following this method, intensity ranges of Cin8 motor fluorescence are assigned as <200, 200-400, and >400 for single Cin8 molecules, pairs of Cin8 molecules (dimer of Cin8 tetramer), and Cin8 oligomers, respectively12.
- Intensity distribution analysis for Cin8-GFP motors
- Measure the mean florescence intensity of all the fluorescent Cin8-GFP motors in the first frame of the time-lapse sequence using TrackMate plugin in ImageJ-Fiji as described in step 4.2.2.
- Plot a histogram of the mean intensities of Cin8-GFP with a bin size of 20 a.u. and fit the major peak of the histogram to a Gaussian curve (Figure 4B).
NOTE: Intensity distribution analysis complements the cluster size determination for Cin8-GFP motors from the photobleaching experiments. The Gaussian curve fitted to the intensity distribution histogram for the Cin8-GFP population peaks at ~125 a.u., which is consistent with the average intensity of single tetrameric Cin8 molecules containing either one, two, three, or four fluorescent (non-bleached) GFP molecules, with each fluorescent GFP molecule contributing ~50 a.u. Thus, using this intensity distribution method, the contribution of one GFP molecule can also be calculated, which can be further utilized to assign the cluster size of Cin8-GFP molecules.
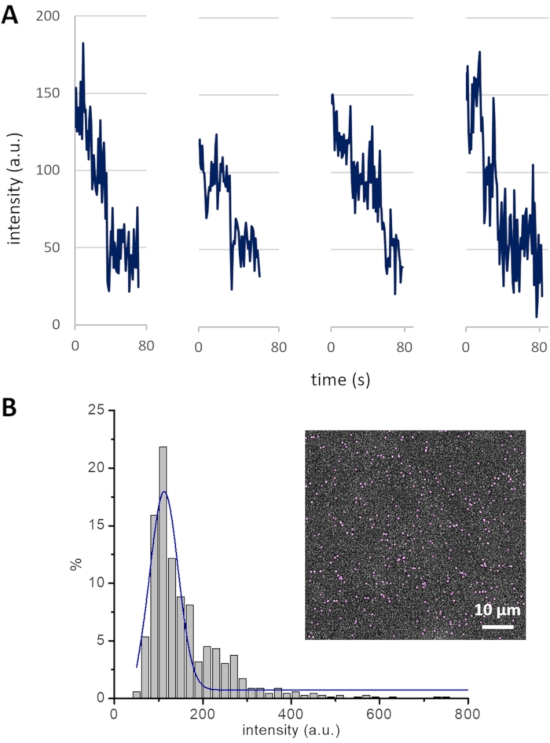
Figure 4: Cin8-GFP bleaching profile and intensity distribution. (A) Photobleaching of GFP in four different Cin8-GFP motors. Single photobleaching steps, each likely representing the photobleaching of one GFP, lead to a drop in fluorescence intensity of ~50 a.u. (B) The intensity distribution of Cin8-GFP motors in the first frame of a time-lapse sequence (inset). The Gaussian peak (blue) centered at ~125 a.u represents single Cin8-GFP molecules. This peak exhibits the average intensity of single Cin8 tetramers with one, two, three, or four fluorescent GFP molecules, with each GFP molecule contributing ~50 a.u. to the total intensity (i.e., (50 + 100 + 150 + 200) / 4 = 125). Please click here to view a larger version of this figure.
- Tracking the Cin8-GFP molecules motility along the MT tracks
- Crop the MT to be analyzed in the time-lapse sequence of recorded frames by highlighting it with the Rectangle tool, and then choosing Image > Crop.
- Choose a fluorescent Cin8-GFP particle for the analysis. Record the particle coordinates in each frame (time point) of the time lapse sequence using the Point Tool and Measure options. Perform similar recording of coordinates for other fluorescent particles in the time-lapse sequence.
- Assign cluster size to all the examined Cin8-GFP particles in the first frame of their appearance, as described in step 4.2.
- Mean displacement (MD) and mean square displacement (MSD) analyses
- From the coordinates of Cin8-GFP movements determined in step 4.4, calculate the displacements of Cin8-GFP at each time point with respect to the initial coordinates, using the equation for calculation of distance between two points with given coordinates:
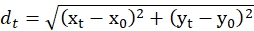
where, dt is the displacements of Cin8-GFP at the time t, xt and yt are the respective coordinates at time t. x0 and y0 are the respective coordinates of Cin8-GFP at t = 0. - Calculate from these displacement values the displacement for all possible time intervals for a specific Cin8-GFP particle. Repeat the procedure for all the examined Cin8-GFP particles.
- Plot the mean displacement (MD) of all the examined Cin8-GFP particles versus time interval and subject to a linear fit, MD = v x t + c. The slope of this fit (v) represents the mean velocity of motile Cin8-GFP particles.
NOTE: In this manner, the average velocity of all Cin8-GFP molecules belonging to each cluster size can be calculated separately characterizing the motility of different cluster sizes. In addition to the MD analysis, mean squared displacement (MSD) analysis can also be performed by squaring the displacement values calculated in steps 4.5.1 and 4.5.2. MSD values are plotted versus time interval and fitted to the polynomial curve MSD = v2 x t2 + 2D x t + c, giving the additional parameter D, which is the diffusion coefficient of Cin8-GFP movement. MD analysis should be performed on polarity marked MTs8,10, whereas for the MSD analysis knowledge of the MT polarity is not necessary.
- From the coordinates of Cin8-GFP movements determined in step 4.4, calculate the displacements of Cin8-GFP at each time point with respect to the initial coordinates, using the equation for calculation of distance between two points with given coordinates:
Representative Results
The experiment aims to investigate the motility characteristics of bi-directional motor protein Cin8 of different cluster sizes on single MTs. Representative motility of Cin8-GFP is also evident from the kymographs in Figure 5A, where the spatial position of the motor over time is shown.
For the analysis of the motile properties of Cin8-GFP, first, the cluster size is assigned (step 4.3) to each MT-attached motile Cin8-GFP particle, and then the position of the examined Cin8 particles is tracked as a function of time (step 4.4). For each cluster size category, >40 trajectories of individual Cin8-GFP were extracted from the recordings (Figure 5B). Using the coordinates obtained from tracking analysis, MD and MSD analysis is performed for each cluster size population separately. The velocities are obtained from linear fits to MD as presented in Figure 5C. It was found that single Cin8-GFP molecules move in a unidirectional, minus-end directed manner with high velocity, whereas the Cin8 clusters exhibit considerably lower velocity with a higher propensity for bi-directional motility (Figure 5B,C).
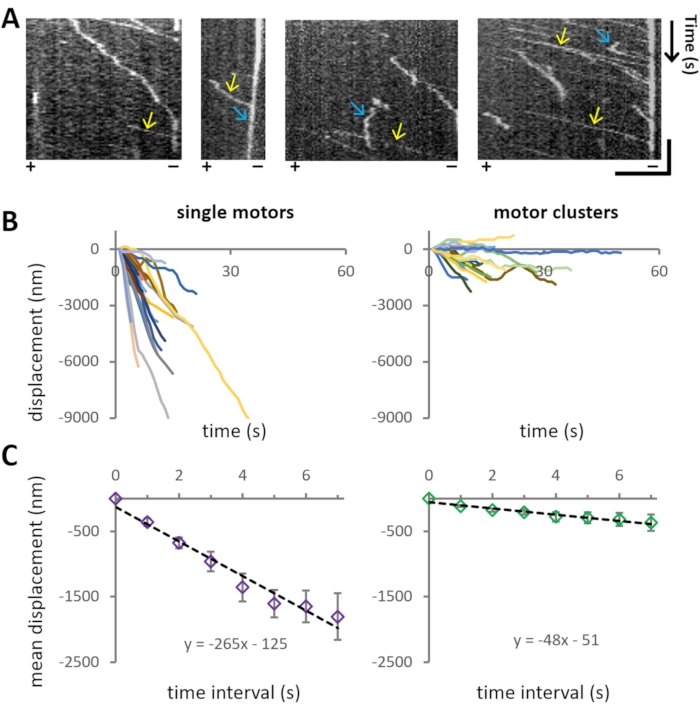
Figure 5: Cin8-GFP motility. (A) Kymographs representing motility of Cin8-GFP motors on MTs. X- and Y-axes represent MT lattice and time, respectively. Yellow arrows mark the fast motility of single Cin8-GFP particles toward the minus-end direction of the MT, whereas blue arrows mark the slow motility of Cin8 clusters in the plus-end direction of the MT. The polarity of the MTs is indicated at the bottom of each kymograph (+/-). Horizontal bar: 4 µm, vertical bar: 20 s. (B) Displacement traces of single motors (left) and clusters (right) of Cin8-GFP motors. The displacement traces were plotted using the coordinates obtained after tracking the individual Cin8-GFP motors as explained in step 4.4. Negative and positive values of displacement indicate movement in the minus-end and plus-end directions of the MT, respectively. Note that under the same assay, the motility of Cin8 clusters is slower and bi-directional compared to the single molecules of Cin8. (C) Plots of mean displacement (MD) ± SEM, of single molecules (left) and clusters (right) of Cin8 motors as a function of the time interval. Black lines represent linear fits of the plot (MD = v xt + c, where v is the mean velocity, t is the time interval and c represents the intercept). From the fitting, it is evident that the mean velocity for single motors and clusters of Cin8 is -265 ± 20 nm/s and -48 ± 5 nm/s, respectively. Please click here to view a larger version of this figure.
Discussion
In this work, a protocol for single-molecule motility assay with the bi-directional kinesin-5 Cin8 and the motility analysis are presented. The full-length Cin818 including the native nuclear localization signal (NLS) at the C-terminal has been purified from the native host S. cerevisiae. As the Cin8 is a nuclear motor protein, grinding the S. cerevisiae cells under liquid nitrogen is found to be the most efficient method for cell lysis. After lysis, by combining metal affinity and size exclusion chromatography, highly pure Cin8 is obtained, which is important for the single-molecule motility assays. It has been previously reported that there are differences between motile properties of Cin8 in crude extracts and purified samples8. In addition, it has also been reported that MT crowding with motor and non-motor proteins affects the directionality of bi-directional kinesin-5 Cut722. Thus, high purity of the motor is required for reliable motility analysis and conclusions regarding wild-type and mutant motor behavior. The techniques described here can be easily adapted to purify other nuclear proteins from the yeast with appropriate buffer adjustments.
Described here is a highly robust and sensitive single-molecule motility assay with GFP-tagged Cin8. The success of this assay relies heavily on the proper MT polymerization and immobilization to the surface. The strong avidin-biotin interaction is utilized to immobilize the MTs to the hydrophobic glass surface, which irreversibly attaches the MTs. On these immobilized MTs using GFP labeled Cin8, Cin8 motility can be reliably tracked11,12,19.
Cin8 is reported to form clusters containing more than one tetrameric motor10,12, with the motility of these clusters being different from that of single Cin8 molecules. To accurately characterize Cin8 motility as a function of its size, a fluorescence intensity-based method has been developed to identify the cluster size of each Cin8 particle12. Based on this size categorization, motility is analyzed separately in each size category. Following this size-based analysis, insightful details are provided, that can be utilized to understand the different behavior of oligomers of the same molecule11,12,19. The cluster size determination procedure described here can be applied to determine the size of a variety of fluorescently labeled molecules. While performing the fluorescence-based size determination, one should be careful to determine the cluster size of Cin8-GFP particles at the first frame of appearance to avoid the impact of bleaching, since the large clusters could appear as smaller ones following photobleaching.
The motility characterization is performed by the MD and/or MSD analyses. If it is of interest to determine only the motor velocity, MD analysis is sufficient. However, if motor motility contains both active and passive components and determination of the diffusion coefficient is also required, MSD analysis should be performed20,23,24,25. For both MD and MSD analyses, the coordinates of the motor for every time point need to be determined. For efficient tracking, it is important to keep the motor concentration optimum. The MTs should not be too crowded with motors; ideally, there should be 3-4 Cin8-GFP motors/particles at a time on an MT of ~10 µm. Automated tools such as the "KymoButler" or "TrackMate" plugin in ImageJ-Fiji can also be used to track the motile motors26,27. These automated tools save time and work, but they have a few limitations. For example, if the motility of some particles is very slow, these tools can read them as non-motile particles. In addition, these tools have limits in recognizing low-intensity molecules. Therefore, they can exhibit a high-intensity bias. On the other hand, manual tracking (although time-consuming) is less sensitive to tracking errors.
In summary, this protocol, starting from the purification of Cin8 overexpressed in S. cerevisiae, explains comprehensively the single-molecule motility assay and the subsequent motility analysis of this bi-directional kinesin-5. This protocol can be followed easily to purify and characterize the motility of motor proteins such as Cin8. Moreover, the different parts of the protocol can be adapted to purify proteins from yeast or develop single-molecule motility assays for different motor proteins and their motility characterization.
Disclosures
The authors have nothing to disclose.
Acknowledgements
This research was supported in part by the Israel Science Foundation grant (ISF-386/18) and the Israel Binational Science Foundation grant (BSF-2019008), awarded to L.G.
Materials
| Adenine | FORMEDIUM | DOC0230 | |
| ATP | Sigma | A7699 | |
| Biotinylated-BSA | Sigma | A8549 | |
| Casein | Sigma | C7078 | |
| Catalase (C40) | Sigma | C40 | |
| Creatine-Kinase | Sigma | C3755 | |
| Dithiothreitol (DTT) | Sigma | D0632 | |
| EDTA | Sigma | E5134 | |
| EGTA | Sigma | E4378 | |
| Fluorescence filter set for GFP | Chroma | 49002: ET-EGFP (FITC/Cy2) | |
| Fluorescence filter set for Rhodamine | Chroma | 49004: ET-CY3/TRITC | |
| Fluorescence inverted microscope | Zeiss | Axiovert 200M | |
| Galactose | Tivan Biotech | GAL02 | |
| Glucose | Sigma | G8270 | |
| Glucose Oxidase | Sigma | G7141 | |
| Glycerol | Sigma | G5516 | |
| GlycylGlycine | Merck | G0674 | |
| GMPCPP | Jana Bioscience | Nu-405L | |
| GTB | Cytoskeleton | BST01-010 | |
| GTP | Sigma | G8877 | |
| Histidine | Duchefa Biochemie | H0710.0100 | |
| ImageJ-FIJI software | https://imagej.net/plugins/trackmate/ | version 2.1.0/1.53c; Java 1.8.0_172 [64-bit] for Windows 10 | |
| Imidazole | Sigma | I0125 | |
| InstantBlue Coomassie Protein Stain | Abcam | ab119211 | |
| Lens | Zeiss | 100x/1.4 oil DIC objective | |
| Lysine | FORMEDIUM | DOC0161 | |
| Magnesium Chloride | Sigma | M8266 | |
| Methionine | Duchefa Biochemie | M0715.0100 | |
| Neo | Andor Technologies | sCMOS camera | |
| NeutraAvidin | Life | A2666 | |
| Ni-NTA Agarose | Invitrogen | R901-15 | |
| Phospho-Creatine | Sigma | P1937 | |
| Pipes | Sigma | P1851 | |
| Pluronic acid F-127 (poloxamer) | Sigma | P2443 | |
| Potassium Chloride | Sigma | P9541 | |
| Raffinose | Tivan Biotech | RAF01 | |
| Size Exclusion chromatography instument | GE Healthcare | AKTA Pure | |
| Spectrophotometer | ThermoFisher Scientific | NanoDrop | |
| Superose-6 10/300 GL | GE Healthcare | 17-5172-01 | |
| Tris | Roshe | 10708976001 | |
| Triton X-100 | Sigma | T8787 | |
| Tryptophan | Duchefa Biochemie | T0720.0100 | |
| Tubulin protein | Cytoskeleton | T240 | |
| Tubulin, biotinylated | Cytoskeleton | T333P | |
| Tubulin, TRITC Rhodamine | Cytoskeleton | TL530M | |
| Uracil | Sigma | U0750-100G | |
| Yeast nitrogen base | FORMEDIUM | CYN0401S | |
| α-GFP antibody | Santa Cruz Biotechnology | SC8036 | |
| β-mercaptoethanol | Sigma | M3148 |
References
- Singh, S. K., Pandey, H., Al-Bassam, J., Gheber, L. Bidirectional motility of kinesin-5 motor proteins: structural determinants, cumulative functions and physiological roles. Cellular and Molecular Life Sciences. 75 (10), 1757-1771 (2018).
- Hoyt, M. A., He, L., Totis, L., Saunders, W. S. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. 유전학. 135 (1), 35-44 (1993).
- Saunders, W. S., Hoyt, M. A. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 70 (3), 451-458 (1992).
- Hoyt, M. A., He, L., Loo, K. K., Saunders, W. S. Two Saccharomyces cerevisiae kinesin-related gene products required for mitotic spindle assembly. Journal of Cell Biology. 118 (1), 109-120 (1992).
- Gerson-Gurwitz, A., et al. Mid-anaphase arrest in S. cerevisiae cells eliminated for the function of Cin8 and dynein. Cellular and Molecular Life Sciences. 66 (2), 301-313 (2009).
- Fridman, V., Gerson-Gurwitz, A., Movshovich, N., Kupiec, M., Gheber, L. Midzone organization restricts interpolar microtubule plus-end dynamics during spindle elongation. EMBO Reports. 10 (4), 387-393 (2009).
- Movshovich, N., et al. Slk19-dependent mid-anaphase pause in kinesin-5-mutated cells. Journal of Cell Science. 121 (15), 2529-2539 (2008).
- Gerson-Gurwitz, A., et al. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. Embo Journal. 30 (24), 4942-4954 (2011).
- Roostalu, J., et al. Directional switching of the kinesin Cin8 through motor coupling. Science. 332 (6025), 94-99 (2011).
- Shapira, O., Goldstein, A., Al-Bassam, J., Gheber, L. A potential physiological role for bi-directional motility and motor clustering of mitotic kinesin-5 Cin8 in yeast mitosis. Journal of Cell Science. 130 (4), 725-734 (2017).
- Goldstein-Levitin, A., Pandey, H., Allhuzaeel, K., Kass, I., Gheber, L. Intracellular functions and motile properties of bi-directional kinesin-5 Cin8 are regulated by neck linker docking. eLife. 10, 71036 (2021).
- Pandey, H., et al. Drag-induced directionality switching of kinesin-5 Cin8 revealed by cluster-motility analysis. Science Advances. 7 (6), 1687 (2021).
- Pandey, H., Popov, M., Goldstein-Levitin, A., Gheber, L. Mechanisms by which kinesin-5 motors perform their multiple intracellular functions. International Journal of Molecular Sciences. 22 (12), 6420 (2021).
- Fridman, V., et al. Kinesin-5 Kip1 is a bi-directional motor that stabilizes microtubules and tracks their plus-ends in vivo. Journal of Cell Science. 126, 4147-4159 (2013).
- Edamatsu, M. Bidirectional motility of the fission yeast kinesin-5, Cut7. Biochemical and Biophysical Research Communications. 446 (1), 231-234 (2014).
- Popchock, A. R., et al. The mitotic kinesin-14 KlpA contains a context-dependent directionality switch. Nature Communications. 8, 13999 (2017).
- Bodrug, T., et al. The kinesin-5 tail domain directly modulates the mechanochemical cycle of the motor domain for anti-parallel microtubule sliding. eLife. 9 (9), 51131 (2020).
- Gheber, L., Kuo, S. C., Hoyt, M. A. Motile properties of the kinesin-related Cin8p spindle motor extracted from Saccharomyces cerevisiae cells. Journal of Biological Chemistry. 274 (14), 9564-9572 (1999).
- Pandey, H., et al. Flexible microtubule anchoring modulates the bi-directional motility of the kinesin-5 Cin8. Cellular and Molecular Life Sciences. 78 (16), 6051-6068 (2021).
- Shapira, O., Gheber, L. Motile properties of the bi-directional kinesin-5 Cin8 are affected by phosphorylation in its motor domain. Scientific Reports. 6, 25597 (2016).
- Schindelin, J., et al. Fiji: an open-source platform for biological-image analysis. Nature Methods. 9 (7), 676-682 (2012).
- Britto, M., et al. Schizosaccharomyces pombe kinesin-5 switches direction using a steric blocking mechanism. Proceedings of the National Academy of Sciences of the United States of America. 113 (47), 7483-7489 (2016).
- Kapitein, L. C., et al. Microtubule cross-linking triggers the directional motility of kinesin-5. Journal of Cell Biology. 182 (3), 421-428 (2008).
- Furuta, K., Edamatsu, M., Maeda, Y., Toyoshima, Y. Y. Diffusion and directed movement in vitro motile properties of fission yeast kinesin-14 Pkl1. Journal of Biological Chemistry. 283 (52), 36465-36473 (2008).
- Katrukha, E. A., et al. Probing cytoskeletal modulation of passive and active intracellular dynamics using nanobody-functionalized quantum dots. Nature Communications. 8, 14772 (2017).
- Tinevez, J. Y., et al. TrackMate: An open and extensible platform for single-particle tracking. Methods. 115, 80-90 (2017).
- Jakobs, M. A. H., Dimitracopoulos, A., Franze, K. KymoBulter, a deep learning software for automated kymograph analysis. eLife. 8, 42288 (2019).

