Measuring the Bending Stiffness of Bacterial Cells Using an Optical Trap
Summary
We present a protocol for bending filamentous bacterial cells attached to a cover-slip surface with an optical trap to measure the cellular bending stiffness.
Abstract
Protocol
- Grow E. coli cells in Luria-Beltrani broth (LB) medium to exponential phase (OD = 0.2-0.4).
- Grow the culture in LB supplemented with 50 μg/mL of cephalexin for 15 min to induce filamentous growth, and then concentrate the culture by 5 times thru centrifugation.
- Make PEI-coated coverslip by flowing 1% polyethylenimine diluted in water into a flow chamber, and wash with water after a 5-minute incubation.
- Flow the concentrated cell culture into the chamber, and washed with a mixture of LB and cephalexin(50μg/mL) after 3 min to remove unattached cells.
- Incubate the chamber at 37°C for 30 min-1 hour to let the attached cells grow before placement on the optical trapping instrument.
- Make polylysine-coated beads by incubating 0.5-μm-diameter polystyrene beads (Bangs Labs) in 0.1% polylysine diluted in water for 30 min. Then wash the beads 3 times and resuspend in water.
- Dilute the bead solution by a factor of two into LB with cephalexin (50μg/mL), and add it into the flow chamber.
- Optically trap a floating bead and touch it to the free tip of a cell. (We use a Mad City Labs piezo stage to control of the motion of the sample.) When a suitable bead/cell combination is found, wash the chamber with cephalexin in LB (50μg/mL) to remove unattached beads.
- Run custom-written LabView program to apply bending forces to cell and record force-displacement data.
Program Details
- Calibrate detector response by raster scanning the attached bead within the detection laser beam and recording 3D PSD voltage signals.
- Identify cell long axis in microscope image.
- Move cell in steps in a direction perpendicular to the cell long axis.
- Record the distance moved and the displacement from the optical trap.
- Convert displacement to applied force using the previously measured trap stiffness and save tip displacement and force to file.
Secrets to Success: In Step 8), one needs to find a cell with a well-defined stuck end. Some cells are just stuck at one tip, and the bending force at the other tip leads to a whole-cell pivoting rather than bending. A suitable pair is found by bending each cell quickly by hand using the joystick-controlled stage motion.
Representative Results:
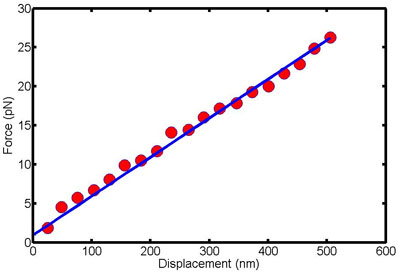
Figure 1. This figure shows force-displacement data for a single cell. The slope of this line is the bending stiffness of the cell.
Discussion
The protocol presented here is designed to quantitatively measure the bending properties of bacterial cells. The experiment setup can be applied to any rod-shaped cell that can be made to grow filamentously. We have successfully used this setup to probe the effects of cytoskeletal filaments on the bending stiffness of E. coli cells. This same technique can be used to evaluate the roles of pressure, cell wall stiffness and other intracellular components in determining overall bending stiffness of cells.
Disclosures
The authors have nothing to disclose.
Acknowledgements
We acknowledge helpful advice from Mingzhai Sun on binding cells to surfaces. We thank Ned Wingreen and Zemer Gitai for valuable discussions. This research was supported by National Institutes of Health grant P50GM07150, National Science Foundation CAREER award PHY-0844466 and the Alfred P. Sloan Foundation.
Materials
| Material Name | Type | Company | Catalogue Number | Comment |
|---|---|---|---|---|
| cephalexin | Sigma | C4895-5G | ||
| polyethylenimine | Sigma | 181978-5G | ||
| polylysine | Sigma | P8920 | ||
| 0.5-μm-diameter polystyrene beads | Bangs Laboratory | PS03N | ||
| Nano-LP Series nanopositioning system | Mad City Labs | NanoLP series | http://www.madcitylabs.com/nanolpseries.html |
References
- Janmey, P. A., McCulloch, C. A. Cell mechanics: integrating cell responses to mechanical stimuli. Annu Rev Biomed Eng. 9, 1-34 (2007).
- Morris, D. M., Jensen, G. J. Toward a biomechanical understanding of whole bacterial cells. Annu Rev Biochem. 77, 583-613 (2008).

