Assessing Cell Cycle Progression of Neural Stem and Progenitor Cells in the Mouse Developing Brain after Genotoxic Stress
Summary
Administration of two analogs of thymidine, EdU and BrdU, in pregnant mice allows the analysis of cell cycle progression in neural and progenitor cells in the embryonic mouse brain. This method is useful to determine the effects of genotoxic stress, including ionizing radiation, during brain development.
Abstract
Neurons of the cerebral cortex are generated during brain development from different types of neural stem and progenitor cells (NSPC), which form a pseudostratified epithelium lining the lateral ventricles of the embryonic brain. Genotoxic stresses, such as ionizing radiation, have highly deleterious effects on the developing brain related to the high sensitivity of NSPC. Elucidation of the cellular and molecular mechanisms involved depends on the characterization of the DNA damage response of these particular types of cells, which requires an accurate method to determine NSPC progression through the cell cycle in the damaged tissue. Here is shown a method based on successive intraperitoneal injections of EdU and BrdU in pregnant mice and further detection of these two thymidine analogues in coronal sections of the embryonic brain. EdU and BrdU are both incorporated in DNA of replicating cells during S phase and are detected by two different techniques (azide or a specific antibody, respectively), which facilitate their simultaneous detection. EdU and BrdU staining are then determined for each NSPC nucleus in function of its distance from the ventricular margin in a standard region of the dorsal telencephalon. Thus this dual labeling technique allows distinguishing cells that progressed through the cell cycle from those that have activated a cell cycle checkpoint leading to cell cycle arrest in response to DNA damage.
An example of experiment is presented, in which EdU was injected before irradiation and BrdU immediately after and analyzes performed within the 4 hr following irradiation. This protocol provides an accurate analysis of the acute DNA damage response of NSPC in function of the phase of the cell cycle at which they have been irradiated. This method is easily transposable to many other systems in order to determine the impact of a particular treatment on cell cycle progression in living tissues.
Introduction
During embryonic brain development, projection neurons of the cerebral cortex are generated in the ventricular zone, a pseudostratified epithelium composed of different types of neural stem and progenitor cells (NSPC) that lines the lateral ventricles. Among NSPC, the radial glial cells (RGC), which serve as neural stem cells, undergo interkinetic nuclear migration (INM): they perform mitosis at the surface of the ventricle and S-phase at the basal limit of the ventricular zone (VZ)1,2,3. They can divide either symmetrically to generate two RGC or asymmetrically to generate one RGC and one neuron or an intermediate progenitor cell (IPC)4,5. IPC migrate to an overlying proliferating layer called the subventricular zone (SVZ), where after one last symmetric division they generate two immature projection neurons6-8. Contrary to RGC, IPC do not undergo INM (reviewed in 9). Newly generated neurons migrate radially along the radial fibers through the intermediate zone (IZ) to reach their final destination in the cortical plate (CP)10,8. The perfect timing of all these events is essential for a correct cortical development. For instance the switch from proliferation to differentiation of neural progenitor populations is controlled by the G1 phase duration11,12. The lengthening of the G1 phase correlates thus with cell differentiation.
Genotoxic stress, such as ionizing radiation, severely impair brain development (reviewed in 13). We and others have shown that NSPC are highly prone to radiation-induced apoptosis14,15,16. Ionizing radiations induce DNA double strand breaks that are the most severe damage to proliferating cells. One essential component of DNA Damage Response (DDR) in cycling cells is the activation of cell cycle checkpoints at the G1/S or G2/M transitions or during S phase (intra-S checkpoint)17-21. They block the cell cycle progression to provide time for DNA damage repair or elimination of too damaged cells. Consequently, cell death as well as delayed cell cycle progression may alter brain development in response to ionizing radiation exposure22-24. It was thus interesting to develop a method to assess the activation of cell cycle checkpoints in NSPC in the irradiated mouse embryonic brain.
The progression of the cell cycle is routinely followed using the incorporation of a thymidine analog, 5-Bromo-2’-deoxyUridine (BrdU). BrdU is incorporated during the S phase of the cell cycle, when DNA is replicating. The use of an antibody against BrdU allows thereafter the detection of cells that were in S phase during the pulse of BrdU.
A novel thymidine analog, 5-Ethynyl-2’-deoxyUridine (EdU) is detected by a fluorescent azide. The different ways to detect EdU and BrdU do not cross-react25, enabling the simultaneous detection of both thymidine analogs, which is useful for the study of cell cycle progression. Usually cells are first pulsed with EdU and then pulsed with BrdU, where the time between both incorporations lasts couple of hours25,26. Addition of BrdU in culture media containing EdU results in preferentially incorporation of BrdU into the DNA with the exclusion of EdU, while simultaneous addition of EdU25 with equimolar or half equimolar BrdU to the media results in only BrdU incorporation27. This simplifies the dual labeling protocol by eliminating the wash steps that are normally required to remove the first label from the culture media prior to addition of the second label. This also is of a particular interest for in vivo study, where the washing steps are not possible, to determine the precise timing of S phase entry or exit of cell population.
Recently, a method of dual-pulse labeling in embryonic mouse brain using EdU and BrdU23,24,22 has been developed to analyze cell cycle progression and INM of NSPC after in utero irradiation. Moreover it has been demonstrated that, during S phase, mouse cells replicate first the euchromatin regions and then the pericentric heterochromatin28,29,30. Interestingly, pericentric heterochromatin of different chromosomes clustered in interphasic nuclei to form heterochromatic foci also known as chromocenters and easily detectable by DAPI staining as bright foci. Therefore, the differential EdU and BrdU stainings of euchromatin and chromocenters helped us to analyze more precisely S phase progression of NSPC.
This method allowed the demonstration of the apparent lack of G1/S checkpoint in NSCP22-24, which is quite surprising since this checkpoint is supposed to be critical for genome stability. Several experimental designs based on various combinations of EdU and BrdU pulses have been used to analyze cell cycle progression in the ventral and dorsal telencephalon. Here, we give an example of protocols allowing the study of the acute DNA damage of NSPC within the first 4 hr following in utero irradiation of E14.5 mouse embryos.
Protocol
1. Animal Procedures
This protocol has been designed in compliance with the European Communities Council Directive of November 24, 1986 (86/609/EEC) and has been approved by our institutional committee on animal welfare (CETEA-CEA DSV IdF).
- Injections with EdU/BrdU and irradiation of E14.5 pregnant mice (Figure 2A).
- Prepare a solution of EdU at 1 mg/ml and BrdU at 5 mg/ml in PBS. Perform an intra-peritoneal injection (IP) of 100 µl of EdU solution using a 25 G needle into pregnant mice. 1.5 hr later, irradiate the mice (total body irradiation) at 0 Gy or 2 Gy (0.6 Gy/min). Immediately after, inject (IP) 200 µl of BrdU solution using a 25 G needle into the pregnant mice.
- Preparation of coronal slices of embryonic brains.
- At 1 hr or 4 hr after irradiation, euthanize the pregnant mice by carbon dioxide.
- Open the abdominal cavity over three centimeters using scissors. Separate the two uterine horns from the rest of the uterus by sectioning both extremities of the horns. Transfer the uterine horns into a Petri dish containing PBS 0.6% glucose. Open the uterine horns with the forceps in order to isolate the embryos. Remove the amniotic sac of each embryo using forceps.
- Dissect embryonic heads with forceps and fix them by immersion in 4% paraformaldehyde (PFA, pH=7.4) O/N at 4 °C.
- Rinse embryonic heads in PBS for at least one night at 4 °C. Heads can be kept in PBS for several days.
- Embed heads in paraffin using a vacuum infiltration processor as indicated in Table 1. Heads embedding can also be performed under a chemical hood for ethanol and xylen baths, and then in a 65 °C oven for the paraffin baths.
- Prepare 5-µm thick coronal sections with a microtome.
- Mount onto polylysine-coated microscope slides.
- Slides can be kept at room temperature (RT) for several months.
2. EdU/BrdU Staining
- Deparaffinization and antigen unmasking
- Deparaffinize the paraffin-embedded brain sections by immersion of slides in 3 baths of toluene for 5 min.
- Rehydrate for 3 min in 2 baths of each solutions of decreasing ethanol concentration as following: ethanol 100%, ethanol 95%, ethanol 70%, and 5 min in H2O. Slides can be kept in deionized water for several hr.
- Boil the slices for 10 min in citrate solution (10 mM, pH 6), and then incubate in deionized water for 5 min.
- EdU staining
- Perform EdU detection according to the manufacturer’s protocol. Permeabilize cells with 0.5% triton X-100 in PBS for 10min.
- Prepare 1X EdU buffer additive by diluting the 10X solution in deionized water. This solution should be freshly made and used on the same day.
- Prepare EdU reaction cocktail, including EdU Alexa Fluor 488 azide. It is important to add the ingredients following the order listed in Table 2, otherwise, the reaction will not proceed optimally. Use the EdU reaction cocktail within 15 min of preparation.
- Remove the permeabilization buffer (step 2.1.1), then wash twice with PBS.
- Add 150 µl of EdU reaction cocktail, put a coverslip, and incubate for 30 min in the dark at RT.
- Remove the EdU reaction cocktail, then wash once with PBS.
- BrdU staining
- Prepare saturation buffer with 7.5% goat serum and 7.5% fetal bovine serum in PBS. Add 150 µl of saturation buffer on brain slices, put a coverslip, and incubate for 1 hr at RT to block non-specific antibody binding.
- Remove the coverslip. Add 150 µl of mouse anti-BrdU primary antibody diluted at 1/300 in saturation buffer, put a coverslip and incubate O/N at 4 °C.
- Remove the coverslip, then wash 3 times in PBS.
- Add 150 µl of goat anti-mouse-Alexa594 secondary antibody diluted at 1/400 in saturation buffer, put a coverslip, and incubate for 1 hr at RT.
- Remove the coverslip, then wash once in PBS.
- Nuclear staining: add 150 µl of 4’,6-diamidino-2-phenylindole (DAPI) at 1 µg/ml in PBS, put a coverslip and incubate for 2 min at RT.
- Mount slides in mounting medium.
3. Analysis
- Examine brain sections under a fluorescence microscope with a 20X objective or a confocal microscope and acquire images in three channels (488 nm, 594 nm and UV) as separates files.
- Stack images with Photoshop software.
- Analyze brain sections in a standard sector of the dorsomedial cerebral wall. This sector is 100 µm in its medial-lateral dimension and is divided into 18 bins (or more) of 10 µm height in its radial dimension using a grid superposed on images (Figure 1) as previously described31.
- Align the grid such as the first bin is at the ventricular surface, with its long axis parallel to the ventricular border. Then number the labeled EdU, BrdU and/or pyknotic nuclei (corresponding to apoptotic nuclei) in each bin. Number a nucleus located on the boundary of two bins in the bin which contains its larger part, or in the lower bin, when the nucleus occupies equal areas within the two bins.
- Statistical analysis.
Analyze at least two cortical slices per embryo. Repeat experiments on at least 3 embryos from different litters. Results should be given as mean ± standard error of the mean (SEM). Statistical analyses are conducted with Graphpad Prism using two-way ANOVA and Bonferroni multiple comparison posthoc tests.
Representative Results
In the experiment described in Figure 2, EdU was administered 1.5 hr before irradiation and BrdU just after irradiation. Four types of cells were then distinguished in the cortical slices prepared at 1 or 4 hr post-irradiation, according to the incorporation of either EdU or BrdU, both or none (Figures 2A and 2B). Importantly, neither EdU nor BrdU incorporation changed the level of radiation-induced apoptosis (data not shown). Moreover, the staining methods allow only the detection of EDU and BrdU incorporated during DNA replication in S-Phase but do not have the sensitivity to detect DNA synthesis that is associated with repair even after irradiation. Indeed, i) neither EdU nor BrdU staining was observed at 1 hr post irradiation (PI) in cells of the cortical plate, where are located postmitotic neurons at G0 phase and ii) the number of EdU(+) and/or BrdU(+) was not increased in irradiated compared to unirradiated brains.
EdU(+)BrdU(+) nuclei
EdU(+)BrdU(+) nuclei correspond to NSPC that were in S phase before and after 0 hr PI. They were thus irradiated in S phase. As shown in Figures 3B and 3C their euchromatin is EdU(+) and may be also BrdU(+). Their chromocenters were either BrdU(+) or BrdU(-) depending on S phase completion at the time of sacrifice. Figure 2B shows that at 1 hr PI, EdU(+)BrdU(+) nuclei were localized in S phase bins (i.e. bins 4 to 10 , where NSPC are known to perform S phase31. Their distribution and numbers were not affected by irradiation. This suggests that radiation did not induce a complete block of DNA synthesis at this time.
Consistently with the basal to apical nuclear migration during G2 phase, most EdU(+)BrdU(+) nuclei of NSPC were found near the ventricle at 4 hr PI in unirradiated brains (Figure 2C). This did not occur in 2 Gy irradiated brains in which nuclei remained in S phase bins, many of them being apoptotic. Altogether this data showed that NSPC irradiated in S phase activated intra-S checkpoints in association with delayed INM and cell death.
EdU(+)BrdU(-) nuclei
EdU(+)BrdU(-) nuclei correspond to cells that were in S phase before and not after irradiation. Chromocenters of these nuclei are EdU(+), indicating that cells were in late S-phase when EdU was injected in mice (Figure 3A). Since G2/M phases of NSPC last 2 hr in the developing mouse brain, EdU(+)BrdU(-) NSPC were in G2 at 0 hr PI. Consistently, most EdU(+)BrdU(-) nuclei were found in the first bin near the ventricle in unirradiated brains at 1 hr PI. This is a consequence of INM of NSPC, after S phase, where many of them formed typical mitotic figures (Figure 2B). At 1 hr PI, the number of EdU(+)BrdU(-) nuclei was significantly reduced at the ventricular surface and no mitotic figure was detectable in irradiated brains. These data clearly show that cells irradiated in G2 activated the G2/M checkpoint within 1 hr PI. Furthermore, most of EdU(+)BrdU(-) of these nuclei were apoptotic at 4 hr PI demonstrating that NSPC are highly radiosensitive during G2 (Figure 2C).
EdU(-)BrdU(-) nuclei
Cells with an EdU(-)BrdU(-) nucleus correspond to either cells in G0 (principally immature neurons), or NSPC that remained in G1 from EdU incorporation to animal sacrifice (Figure 3E). Comparison between 1 hr and 4 hr in Figure 2C shows interkinetic nuclear migration making that nuclei that were close to the ventricle in G1 at 1 hr underwent their apical to basal migration at 4 hr, whereas nuclei that were in S phase went toward the ventricular surface.
As shown in Figure 2C many of these nuclei were apoptotic at 4 hr PI. The number of apoptotic EdU(-)BrdU(-) nuclei was maximal near the ventricular surface and decreases in upper bins. These data suggest therefore that radiation-induced apoptosis occurred in NSPC that have been irradiated during early G1 phase and/or post-mitotic migrating neurons. By contrast, none or very few apoptotic nuclei were found in the cortical plate. This is in accordance with the radiation resistance of neurons that have reached their final destination.
EdU(-)BrdU(+) nuclei
EdU(-)BrdU(+) nuclei correspond to cells that entered S phase after irradiation. Typically at 1 hr PI, they showed a diffuse BrdU labelling of euchromatin, but not of chromocenters. Thus they were in early S phase at that time (Figure 3D). At 1 hr PI the number and localization within S phase bins of EdU(-)BrdU(+) nuclei is similar between controls and irradiated brains. Thus S-phase entry during the 1st hr PI was not affected by radiation in the mouse developing brain (Figure 2B). Interestingly, comparison of 1 hr and 4 hr PI in irradiated brains shows a decline in the number of EdU(-)BrdU(+) nuclei and most of these nuclei were apoptotic at 4 hr PI. This suggests that cells that entered S phase after radiation activated intra-S checkpoints and underwent apoptosis thereafter (Figure 2C).

Figure 1. Analysis of Brain section Coronal section of the cerebral hemisphere of E14.5 mouse embryos stained with Dapi. EdU and BrdU staining as well as nuclear morphology are analyzed in a standard sector of the dorsomedial cerebral wall using a grid divided into 18 bins (or more) as described in the text. Scale bar: 20 µm. Please click here to view a larger version of this figure.
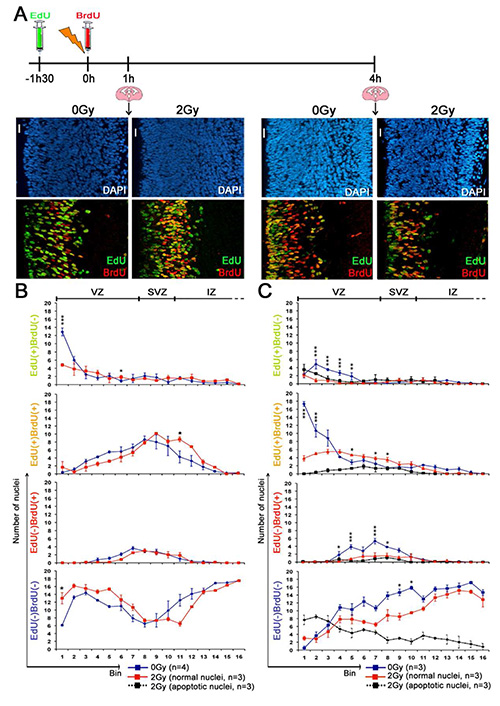
Figure 2. Cell cycle progression of irradiated neural stem and progenitor cell (Figure from Roque et al., Stem cells, 2012). (A) Schematic diagram of the experimental design and DAPI (blue), EdU (green), and BrdU (red) staining patterns found in coronal cortical slices at 1 and 4 hr PI (2 Gy). Scale bar = 20 μm. (B, C) Numbers/bin of EdU(+)BrdU(−), EdU(+)BrdU(+), EdU(−)BrdU(+), or EdU(−)BrdU(−) nuclei with a normal morphology in nonirradiated controls (blue) and with a normal (red) or apoptotic (pyknotic, black) morphology in irradiated (2 Gy) cortices at 1 hr (B) or 4 hr PI (C). No apoptotic nuclei were detected at 1 hr PI (B). Statistical analysis was performed using Bonferroni post hoc tests. Abbreviations: BrdU, 5-bromo-2′-deoxyuridine; DAPI, 4′-6-diamidino-2-phenylindole; EdU, 5-ethynyl-2′deoxyuridine; IZ, intermediate zone; SVZ, subventricular zone; VZ, ventricular zone. Please click here to view a larger version of this figure.
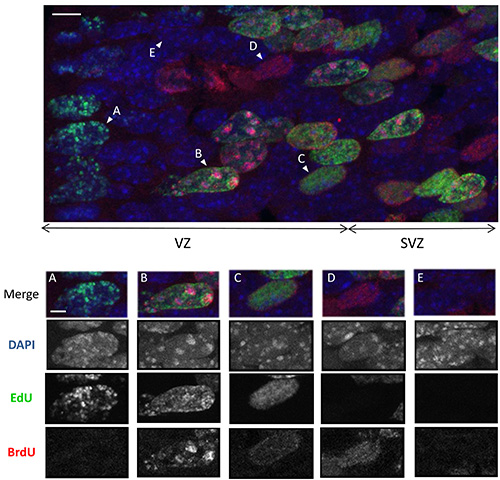
Figure 3. Differential EdU and BrdU staining of euchromatin and chromocenters Upper panel: Z-stack confocal images of EdU (green), BrdU (red) staining and DNA counterstained with DAPI (blue) allowing the detection of chromocenters as bright blue foci. Stainings were performed on an embryonic brain section collected at 1 hr PI in experiments where EdU was delivered 1.5 hr before irradiation and BrdU 0 hr PI (only the unirradiated control is shown) (Scale bar: 5 µm). Please click here to view a larger version of this figure.
Bottom panels show examples of the 5 different patterns of EdU and BrdU staining detected in coronal brain slices. Merge image and single channels are shown (Scale bar: 2 µm):
A) Nucleus with EdU(+) chromocenters and EdU(+) or EdU(-) euchromatin. EdU has been thus incorporated in late S phase during the replication of pericentric heterochromatin: the cells were in G2 at 0 hr PI.
B) Nucleus with EdU(+) euchromatin and BrdU(+) chomocenters: EdU has been incorporated in early S phase when most of euchromatin is replicated and BrdU has been incorporated in late S phase: the cells were in S phase at 0 hr PI.
C) Nucleus with EdU(+)BrdU(+) euchromatin and EdU(-)BrdU(-) chromocenters. EdU and BrdU have been incorporated in the first part of the S phase: The cells were in early S phase at 0 hr PI.
D) Nucleus with BrdU(+) euchromatin and EdU(-)BrdU(-) chromocenter. BrdU has been incorporated in the beginning of the S phase. The cells were in late G1 at 0 hr PI and entered S phase thereafter.
E) Nucleus with EdU(-)BrdU(-) euchromatin and chromocenters. The cells were in G0 (immature neurons) or in G1 phase and did not enter S phase during EdU and BrdU pulses.
| Solution | Incubation | Temperature | Pressure | Agitation | |
| 1 | Alcohol 70% | 30 min | 35 °C | on | on |
| 2 | alcohol 95% | 15 min | 35 °C | on | on |
| 3 | alcohol 95% | 30 min | 35 °C | on | on |
| 4 | alcohol 95% | 45 min | 35 °C | on | on |
| 5 | alcohol 100% | 15 min | 35 °C | on | on |
| 6 | alcohol 100% | 30 min | 35 °C | on | on |
| 7 | alcohol 100% | 1 hr | 35 °C | on | on |
| 8 | Toluene 100% | 30 min | 35 °C | on | on |
| 9 | Toluene 100% | 45 min | 35 °C | on | on |
| 10 | Toluene 100% | 1 hr | 35 °C | on | on |
| 11 | Paraffin | 30 min | 58 °C | on | on |
| 12 | Paraffin | 1 hr | 58 °C | on | on |
| 13 | Paraffin | 1 hr | 58 °C | on | on |
| 14 | Paraffin | 1.5 hr | 58 °C | on | on |
Table 1. Vacuum infiltration processor program for paraffin embedding.
| Reaction components for 1 slide | |
| 1X EdU reaction buffer | 128.64 µl |
| CuSO4 | 6 µl |
| EdU Alexa Fluor azide | 0.36 µl |
| 1X EdU Reaction buffer additive | 15 µl |
| Total volume | 150 µl |
Table 2. EdU reaction cocktails.
Discussion
The experimental design described here based on incorporation of EdU 1.5 hr before irradiation and incorporation of BrdU immediately after irradiation allowed the demonstration that NSPCs are able to activate S and G2/M checkpoints but not the G1/S checkpoint during the 1st hr after a genotoxic stress in the fetal mouse brain. We performed other experiments in which EdU has been injected at different times after irradiation and BrdU, just 1 hr before mice sacrifices, allowing us to specifically appreciate the rate of S phase entry. These experiments have demonstrated that NSPC do not activate a G1/S arrest after a genotoxic stress in vivo22,23.
Therefore, the choice between DNA repair and apoptosis does not occur at the G1/S checkpoint raising the question on the nature of the mechanisms guarantying genome stability in these cells.
The protocol is easy and fast. The different steps present no obvious difficulty. This technique is an alternative to the use of chloro-deoxyuridine (CldU) and iodo-deoxyuridine (IdU) which have been shown to cross-react32.
A broad panel of applications may be considered, as the protocol is easily flexible and adaptable to several biological questions. Examples of changes are proposed in Roque and collaborators23, Etienne and collaborators22, and in Rousseau and collaborators24. It could be also combined with immunofluorescence detection of several cell markers or proteins involved in DNA damage response for instance. Note, that in this case, both BrdU and EdU detection could be performed with different fluorophores that are commercialized, to favor immune detection of other markers.
EdU and BrdU in vivo incorporation in NSPC could also be used in cell sorting by flow cytometry for further biochemical analysis. Finally, this method could be performed to monitor cell cycle progression of NSPC in different types of mutant mice or after other types of genotoxic stresses (other than ionizing radiation) or any other treatment affecting cell cycle progression. Moreover it could be easily adapted to other replicating tissues.
Declarações
The authors have nothing to disclose.
Acknowledgements
The research leading to these results has received funding from the European Union Seventh Framework Programme (FP7/2007-2013) under grant agreement n° 323267, from Electricité de France (EDF) and from l’Agence Nationale de la Recherche – Santé-Environnement et Santé-Travail (ANR-SEST, Neurorad).
Materials
| EdU | Life Technologies | A10044 | 1 mg/ml |
| BrdU | Life Technologies | B5002 | 5 mg/ml |
| IBL 637 | CIS BIO International | ||
| Tissu Tek VIP | Leica | ||
| Microtome | Leica | RM 2125 RT | |
| Triton X-100 | Sigma-Aldrich | 93443 | |
| Click-iT EdU Alexa 488 imaging kit | Life Technologies | C10083 | |
| Anti-BrdU | GE Healthcare | RPN202 | 1/300 |
| Goat anti mouse-Alex594 | Life Technologies | A11001 | 1/400 |
| Fluoromount | SouthernBiotech | 0100-01 | |
| Polysine slide | Thermo scientific | J2800AMNZ | |
| Paraformaldehyde | Sigma-Aldrich | P6148 | |
| PBS | Life Technologies | 20012-068 | |
| DAPI | Sigma-Aldrich | D9542 | |
| Microscope BX51 | Olympus | ||
| Confocal microscope SPE | Leica | ||
| Prism software | Graphpad | Version 5.0c | |
| Photoshop software | Adobe |
Referências
- Rakic, P. Specification of cerebral cortical areas. Science. 241, 170-176 (1988).
- Sauer, F. Mitosis in he neural tube. J Comp Neurol. , 377-399 (1935).
- Taverna, E., Huttner, W. B. Neural progenitor nuclei IN motion. Neuron. 67, 906-914 (2010).
- Committee, B. Embryonic vertebrate central nervous system: revised terminology. Anat Rec. 166, 257-261 (1970).
- Bystron, I., Blakemore, C., Rakic, P. Development of the human cerebral cortex: Boulder Committee revisited. Nature Reviews. Neuroscience. 9, 110-122 (2008).
- Miyata, T., et al. Asymmetric production of surface-dividing and non-surface-dividing cortical progenitor cells. Development. 131, 3133-3145 (2004).
- Haubensak, W., Attardo, A., Denk, W., Huttner, W. B. Neurons arise in the basal neuroepithelium of the early mammalian telencephalon: a major site of neurogenesis. Proc Natl Acad Sci U S A. 101, 3196-3201 (2004).
- Noctor, S. C., Martinez-Cerdeno, V., Ivic, L., Kriegstein, A. R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat Neurosci. 7, 136-144 (2004).
- Merot, Y., Retaux, S., Heng, J. I. Molecular mechanisms of projection neuron production and maturation in the developing cerebral cortex. Seminars in Cell & Developmental Biology. 20, 726-734 (2009).
- Kriegstein, A. R., Noctor, S. C. Patterns of neuronal migration in the embryonic cortex. Trends Neurosci. 27, 392-399 (2004).
- Salomoni, P., Calegari, F. Cell cycle control of mammalian neural stem cells: putting a speed limit on G1. Trends Cell Biol. 20, 233-243 (2010).
- Calegari, F., Haubensak, W., Haffner, C., Huttner, W. B. Selective lengthening of the cell cycle in the neurogenic subpopulation of neural progenitor cells during mouse brain development. J Neurosci. 25, 6533-6538 (2005).
- Andres-Mach, M., Rola, R., Fike, J. R. Radiation effects on neural precursor cells in the dentate gyrus. Cell Tissue Res. 331, 251-262 (2008).
- Semont, A., et al. Involvement of p53 and Fas/CD95 in murine neural progenitor cell response to ionizing irradiation. Oncogene. 23, 8497-8508 (2004).
- Nowak, E., et al. Radiation-induced H2AX phosphorylation and neural precursor apoptosis in the developing brain of mice. Radiat Res. 165, 155-164 (2006).
- Gatz, S. A., et al. Requirement for DNA ligase IV during embryonic neuronal development. J Neurosci. 31, 10088-10100 (2011).
- Denekamp, J. Cell kinetics and radiation biology. International Journal of Radiation Biology and Related Studies in Physics, Chemistry, and Medicine. 49, 357-380 (1986).
- Hartwell, L. H., Weinert, T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 246, 629-634 (1989).
- Weinert, T. A., Hartwell, L. H. Characterization of RAD9 of Saccharomyces cerevisiae and evidence that its function acts posttranslationally in cell cycle arrest after DNA damage. Mol Cell Biol. 10, 6554-6564 (1990).
- Tolmach, L. J., Jones, R. W., Busse, P. M. The action of caffeine on X-irradiated HeLa cells. I. Delayed inhibition of DNA synthesis. Radiat Res. 71, 653-665 (1977).
- Painter, R. B., Young, B. R. Radiosensitivity in ataxia-telangiectasia: a new explanation. Proc Natl Acad Sci U S A. 77, 7315-7317 (1980).
- Etienne, O., Roque, T., Haton, C., Boussin, F. D. Variation of radiation-sensitivity of neural stem and progenitor cell populations within the developing mouse brain. Int J Radiat Biol. 88, 694-702 (2012).
- Roque, T., et al. Lack of a p21waf1/cip -dependent G1/S checkpoint in neural stem and progenitor cells after DNA damage in vivo. Stem Cells. 30, 537-547 (2012).
- Rousseau, L., et al. In vivo importance of homologous recombination DNA repair for mouse neural stem and progenitor cells. PLoS One. 7, 12 (2012).
- Bradford, J. A., Clarke, S. T. Dual-pulse labeling using 5-ethynyl-2′-deoxyuridine (EdU) and 5-bromo-2′-deoxyuridine (BrdU) in flow cytometry. Curr Protoc Cytom. , (2011).
- Deckbar, D., et al. The limitations of the G1-S checkpoint. Cancer Res. 70, 4412-4421 (2010).
- Manual, Click-iT EdU Imaging Kit. , (2011).
- Weidtkamp-Peters, S., Rahn, H. P., Cardoso, M. C., Hemmerich, P. Replication of centromeric heterochromatin in mouse fibroblasts takes place in early, middle, and late S phase. Histochem Cell Biol. 125, 91-102 (2006).
- Wu, R., Terry, A. V., Singh, P. B., Gilbert, D. M. Differential subnuclear localization and replication timing of histone H3 lysine 9 methylation states. Mol Biol Cell. 16, 2872-2881 (2005).
- Wu, R., Singh, P. B., Gilbert, D. M. Uncoupling global and fine-tuning replication timing determinants for mouse pericentric heterochromatin. J Cell Biol. 174, 185-194 (2006).
- Takahashi, T., Nowakowski, R. S., Caviness Jr, V. S. The cell cycle of the pseudostratified ventricular epithelium of the embryonic murine cerebral wall. J Neurosci. 15, 6046-6057 (1995).
- Tuttle, A. H., et al. Immunofluorescent detection of two thymidine analogues (CldU and IdU) in primary tissue. J Vis Exp. , (2010).

