Pooled CRISPR-Based Genetic Screens in Mammalian Cells
Summary
CRISPR-Cas9 technology provides an efficient method to precisely edit the mammalian genome in any cell type and represents a novel means to perform genome-wide genetic screens. A detailed protocol discussing the steps required for the successful performance of pooled genome-wide CRISPR-Cas9 screens is provided here.
Abstract
Genome editing using the CRISPR-Cas system has vastly advanced the ability to precisely edit the genomes of various organisms. In the context of mammalian cells, this technology represents a novel means to perform genome-wide genetic screens for functional genomics studies. Libraries of guide RNAs (sgRNA) targeting all open reading frames permit the facile generation of thousands of genetic perturbations in a single pool of cells that can be screened for specific phenotypes to implicate gene function and cellular processes in an unbiased and systematic way. CRISPR-Cas screens provide researchers with a simple, efficient, and inexpensive method to uncover the genetic blueprints for cellular phenotypes. Furthermore, differential analysis of screens performed in various cell lines and from different cancer types can identify genes that are contextually essential in tumor cells, revealing potential targets for specific anticancer therapies. Performing genome-wide screens in human cells can be daunting, as this involves the handling of tens of millions of cells and requires analysis of large sets of data. The details of these screens, such as cell line characterization, CRISPR library considerations, and understanding the limitations and capabilities of CRISPR technology during analysis, are often overlooked. Provided here is a detailed protocol for the successful performance of pooled genome-wide CRISPR-Cas9 based screens.
Introduction
CRISPR-Cas, short for clustered regularly interspaced short palindromic repeats and CRISPR-associated nuclease, consists of a single nuclease protein (e.g., Cas9) in complex with a synthetic guide RNA (sgRNA). This ribonucleoprotein complex targets the Cas9 enzyme to induce double-stranded DNA breaks at a specific genomic locus1. Double-stranded breaks can be repaired via homology directed repair (HDR) or, more commonly, through non-homologous end joining (NHEJ), an error prone repair mechanism that results in insertion and/or deletions (INDELS) that frequently disrupt gene function1. The efficiency and simplicity of CRISPR enables a previously unattainable level of genomic targeting that far surpasses previous genome editing technologies [i.e., zinc finger nucleases (ZNF) or transcription activator-like effector nucleases (TALENS), both of which suffer from heightened design complexity, lower transfection efficiency, and limitations in multiplex gene editing2].
The basic research application of CRISPR single-guide RNA-based genome editing has allowed scientists to efficiently and inexpensively interrogate the functions of individual genes and topology of genetic interaction networks. The ability to perform functional genome-wide screens has been greatly enhanced by use of the CRISPR-Cas system, particularly when compared to earlier genetic perturbation technologies such as RNA interference (RNAi) and gene trap mutagenesis. In particular, RNAi suffers from high off-target effects and incomplete knockdown, resulting in lower sensitivity and specificity compared to CRISPR3,4,5, while gene trap methods are only feasible in haploid cells for loss-of-function screens, limiting the scope of cell models that can be interrogated6. The ability of CRISPR to generate complete gene knock-out provides a more biologically robust system to interrogate mutant phenotypes, with low noise, minimal off-target effects and consistent activity across reagents5. CRISPR-Cas9 sgRNA libraries that target the entire human genome are now widely available, allowing simultaneous generation of thousands of gene knock-outs in a single experiment3,7,8,9.
We have developed unique CRISPR-Cas9 genome-wide sgRNA lentiviral libraries called the Toronto Knock-out (TKO) libraries (available through Addgene) that are compact and sequence-optimized to facilitate high resolution functional genomics screens. The latest library, TKOv3, targets ~18,000 human protein-coding genes with 71,090 guides optimized for editing efficiency using empirical data10. Additionally, TKOv3 is available as a one-component library (LCV2::TKOv3, Addgene ID #90294) expressing Cas9 and sgRNAs on a single vector, alleviating the need to generate stable Cas9-expressing cells, enabling genome-wide knock-out across a broad range of mammalian cell types. TKOv3 is also available in a vector without Cas9 (pLCKO2::TKOv3, Addgene ID# 125517) and can be utilized in cells that express Cas911.
A genome-wide CRISPR-Cas9 edited cell population can be exposed to different growth conditions, with the abundance of sgRNAs over time quantified by next-generation sequencing, providing a readout to assess drop-out or enrichment of cells with traceable genetic perturbations. CRISPR knock-out libraries can be harnessed to identify genes that, upon perturbation, cause cellular fitness defects, moderate drug sensitivity (e.g., sensitive or resistant genes), regulate protein expression (e.g., reporter), or are required for a certain pathway function and cellular state12,13,14. For example, differential fitness screens in a cancer cell line can identify both depletion or reduction of oncogenes and enrichment or an increase of tumor suppressors genes3,14,15. Similarly, using intermediate doses of therapeutic drugs can reveal both drug resistance and sensitization genes16,17.
Provided here is a detailed screening protocol for genome-scale CRISPR-Cas9 loss-of-function screening using the Toronto Knock-out libraries (TKOv1 or v3) in mammalian cells from library generation, screening performance to data analysis. Although this protocol has been optimized for screening using the Toronto Knock-out libraries, it can be applied and become scalable to all CRISPR sgRNA pooled libraries.
Protocol
The experiments outlined below should follow the institute’s Environmental Health and Safety Office guidelines.
1. Pooled CRISPR sgRNA lentiviral library plasmid amplification
- Dilute the ready-made CRISPR sgRNA plasmid DNA library to 50 ng/μL in TE (e.g., TKOv3).
- Electroporate the library using electrocompetent cells. Set up a total of four electroporation reactions as described below.
- Add 2 μL of 50 ng/μL TKO library to 25 μL of thawed electrocompetent cells to pre-chilled cuvettes (1.0 mm) on ice.
- Electroporate using optimal settings suggested by the manufacturer’s protocol. Within 10 s of the pulse, add 975 μL of Recovery Medium (or SOC medium) to the cuvette.
- Transfer electroporated cells to a culture tube and add 1 mL of Recovery Medium. Incubate tubes in a shaking incubator at 250 rpm for 1 h at 37 °C.
- Set up a dilution plate to titer the library and estimate transformation efficiency.
- Pool all 8 mL of recovered cells and mix well. Transfer 10 μL of the pooled cells to 990 μL of Recovery Medium for an 800-fold dilution and mix well.
- Plate 20 μL of the dilution onto a pre-warmed 10 cm LB + carbenicillin (100 µg/L) agar plate. This results in a 40,000-fold dilution of the transformants that will be used to calculate the transformation efficiency.
- Plate 400 μL of recovered cells on each plate across a total of 20 pre-warmed 15 cm LB + carbenicillin agar plates. Incubate the plates for 14–16 h at 30 °C.
NOTE: Growth at this lower temperature minimizes the recombination between long-terminal repeats (LTR)18. - To calculate the transformation efficiency, count the number of colonies on the 40,000-fold dilution plate (step 1.3.2). Multiply the number of colonies counted by 40,000 to obtain the total number of colonies on all plates. Proceed if the total number of colonies represents a library coverage equivalent to minimum of 200x colonies per sgRNA (most optimal is 500-1000x).
- For example, the minimal colony number for TKOv3 library (71,090 sgRNA) is 1.4 x 107, which is equivalent to 200x colonies per sgRNA. If colony representation is insufficient, increase the number of electroporations in step 1.2 based on the number of colonies on the dilution plate to achieve the minimum library coverage.
- Harvest the colonies as described below
- To each 15 cm plate, add 7 mL of LB + carbenicillin (100 µg/L) medium, then scrape the colonies off with a cell spreader. With a 10 mL pipette, transfer the scraped cells into a sterile 1 L conical flask or bottle.
- Once again rinse the plate with 5 mL of LB + carbenicillin medium and transfer the solution to the bottle.
- Repeat for all plates to pool cells from 20 plates into a sterile bottle.
- Mix collected cells with a stir bar for 1 h at room temperature (RT) to break up cell clumps. Transfer cells to pre-weighed centrifuge bottles and centrifuge at 7,000 x g to pellet bacteria, then discard media.
- Weigh the wet cell pellet and subtract the weight of the centrifuge bottle to determine the final weight of the wet pellet. Purify plasmid DNA using a maxi- or mega-scale plasmid purification kit depending on the amount of bacterial pellet each column can process.
2. Large-scale CRISPR sgRNA library lentivirus production
NOTE: All steps in this section of the protocol are performed in a BSL2+ facility in a Class II, Type A2 biosafety cabinet.
- Calculate the number of 15 cm plates required for virus production based on the estimate that 18 mL of virus is typically harvested from one 15 cm plate.
- Prepare cells for transfection by seeding HEK-293T packaging cells in low-antibiotic growth media (DMEM + 10% FBS + optional: 0.1x pen/strep) at 8 x 106 cells per 15 cm plate in 20 mL of media. Incubate cells overnight at 37 °C, 5% CO2. Ensure that the plated cells are 70%–80% confluent and evenly spread at moment of transfection.
- Next day, prepare three transfection plasmids mixture as outlined in Table 1 for 15 cm plates. Calculate the amount of plasmid needed for one transfection and make a mix of plasmids for the number of plates, plus one to be transfected.
- Prepare a lipid-based transfection reagent for each transfection as outlined in Table 2. Aliquot reduced serum media into individual 1.5 mL microcentrifuge tubes for the number of plates to be transfected. Add transfection reagent, mix gently, and incubate for 5 min at RT.
- Following 5 min incubation, add the amount of DNA required for one transfection to the transfection reagent for a 3:1 ratio of transfection reagent-to-µg of DNA complex. Mix gently and incubate for 30 min at RT.
NOTE: Subsequent transfections can be prepared in sets of five or less, with 5 min intervals to optimize for time and avoid over-incubation. - After 30 min of incubation, carefully transfer each transfection mix to each plate of packaging cells. Add the entire mix using a 1 mL pipette tip dropwise in a circular, zigzag motion without disturbing the cell monolayer. Incubate cells at 37 °C for 18 h at 5% CO2.
- Prepare viral harvest media: 500 mL of DMEM medium + 32 mL of BSA stock (20 g/100 mL, dissolved in DMEM, filter sterilized with 0.22 µm filter) + 5 mL of 100x pen/strep.
- After 18 h, remove media (use proper handling of lentivirus waste such as incubation in 1% sodium hypochlorite for 30 min before disposal). Gently replace with 18 mL of viral harvest media to each plate. Incubate cells at 37 °C for 18 h at 5% CO2.
- After 24 h, check packaging cells for abnormal and fused morphology as an indication of good virus production. Then, harvest the lentivirus by collecting all supernatant and transferring into a sterile conical centrifuge tube.
- Spin the media containing virus at 300 x g for 5 min and pellet the packaging cells. Aliquot the supernatant into a sterile polypropylene tube without disturbing the pellet.
- Store the virus at 4 °C for short periods (less than 1 week) or immediately at -80 °C for long-term storage. Aliquot large-scale virus preps to single use volumes for long-term storage to avoid freeze/thawing.
3. Cell line characterization for screening
- Select the desired cell line.
- Measure and record the approximate doubling time of the cells.
- Determine optimal cell plating density for culturing cells every 3–4 cell doublings in a tissue culture vessel of choice (e.g., 15 cm tissue culture plates).
- Determine the puromycin concentration to use in the desired cell line for selection of TKO libraries containing puromycin resistance marker as follows:
- Seed cells in a 12 well plate at the density required to reach confluence after 72 h, then incubate overnight (37 °C, 5% CO2).
- The next day, change to a media containing a dilution range of puromycin concentrations from 0 μg/mL to 10 μg/mL, in 0.5 μg/mL increments. Incubate the cells for 48 h.
- After 48 h, measure the cell viability by cell counting or alamarBlue staining.
- Determine the lowest concentration that kills 100% of cells in 48 h. Use this concentration to select for CRISPR library transduced cell populations in steps 4.6 and 5.2.6.
NOTE: For cell lines with longer doubling times, longer incubations with puromycin can be tolerated. In these situations, determine the kill curve for the incubation time required for <3 cell doublings. Minimize the time for selection to avoid dropout of essential genes before the start of screening.
- Check cells for sensitivity to hexadimethrine bromide (up to 8 μg/mL) by performing a dose response curve in the same method as used for measuring puromycin sensitivity (step 3.2). If toxicity is observed with <8 μg/mL of hexadimethrine bromide, do not use.
4. Functional titration of pooled CRISPR lentivirus library for determination of MOI
- Thaw a fresh aliquot of pooled CRISPR sgRNA library lentivirus (e.g., LCV2::TKOv3) and keep on ice.
- Design a series of virus volumes to test between the range of 0–2 mL (i.e., 0 mL, 0.25 mL, 0.5 mL, 1 mL, and 2 mL).
- Harvest target cells and seed cells in 15 cm plates at the density required to reach confluence in 72 h.
- For each virus volume to be tested, prepare duplicate plates. Add cells, virus, hexadimethrine bromide (8 µg/mL), and media to a final volume of 20 mL. Mix plates thoroughly, sit plates level in incubator and incubate for 24 h (37 °C, 5% CO2).
- After 24 h, remove virus containing media and dispose (use biosafety precautions for handling of lentivirus waste). Optionally, gently wash the plate with warm PBS to remove extraneous virus.
- For each virus condition, replace with 20 mL of media containing puromycin using the concentration determined to kill cells in section 3, to one replicate plate. To the other plate, add 20 mL of fresh media without puromycin. Incubate for 48 h (37 °C, 5% CO2).
- After 48 h, check that all uninfected cells (0 mL virus condition) treated with puromycin are dead. Harvest all plates individually and disperse cells by repeated gentle pipetting.
- Count cells from all the plates and calculate the MOI for each virus volume by comparing cell counts with puromycin selection to cell counts without puromycin (i.e., +/- puromycin).
- Graph results to determine the virus volume that leads to 30%–40% cell survival with puromycin selection versus without puromycin. Use this virus volume to achieve a MOI of 0.3–0.4 during the screen under the same tissue culture conditions.
5. Primary screen infection, selection, and cell passaging
- Select the CRISPR sgRNA library coverage to be maintained throughout the screen (recommended minimum of 200-fold).
- Based on the library coverage, determine the number of cells required to maintain this coverage per sgRNA and the number of cells required for infection at MOI 0.3 (Table 3).
- Determine the number of plates required to set up the infection (Table 4).
- Infecting the cells with CRISPR library
- Harvest cells and seed the required cell number to each 15 cm plate.
- Add hexadimethrine bromide (8 µg/mL) to all plates.
- Add the virus at the volume required for MOI 0.3 to screening and the Control 2 plates. For the Control 1, do not add virus, and replace that volume with media.
- Mix plates thoroughly by tilting. Place plates in incubator, making sure they are level.
NOTE: Batch infections can be done by combining a master mix of virus, media, and hexadimethrine bromide to cells in suspension before plating. - Remove media and replace with fresh media containing puromycin at the concentration determined in step 3.2.4 to the screening and control 1 plates 24 h after virus infection. Add fresh media with no puromycin to the control 2 plate. Incubate cells for 48 h (37 °C, 5% CO2).
- 48 h after puromycin addition, ensure that all uninfected cells are dead (control 1) to confirm puromycin activity, then harvest the infected cells.
- Harvesting infected cell population and cell passaging
- Harvest the puromycin-selected cells from all screening plates into one sterile container. Collect the cells from each control plate separately. Disperse cells by gentle repeated pipetting.
- Count cells from pooled screening cells, control 1, and control 2 separately and calculate the number of cells per 1 mL.
- Calculate MOI and fold coverage achieved as follows:
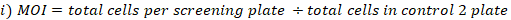
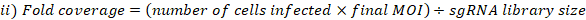
- Collect three replicates of cell pellets from the pooled cells at the selected library coverage for genomic DNA extraction. Centrifuge the cells at 500 x g for 5 min. Wash with PBS. Label the tubes and freeze-dry the cell pellets at -80 °C (these are T0 reference samples).
- Split the pool of infected cells into three replicate groups (e.g., replicate A, replicate B, replicate C), while maintaining library coverage within each replicate. Seed cells at the same seeding density as would normally be used when expanding them. Use the same number of cells for each replicate plate and same total number of cells between replicates.
- Continue to passage cells and harvest three replicates of cell pellets from each replicate of pooled-infected cells as above, every 3–8 days depending on the cell line, for up to 15–20 cell doublings. At each passage, harvest the cells from all plates in each replicate group with each other (i.e., all cells from replicate A plates are re-mixed together, all cells from replicate B plates are re-mixed together, etc.).
- Label each pellet with a time (T) and replicate designation. This corresponds to the number of days post-T0 the pellet is collected (e.g., T3_A, T3_B, T3_C, etc).
- For the negative selection drug screens, allow cells to recover for at least one passage after T0 before treatment. At T3 or T6, split the cells from each replicate group (A, B, C) into drug treatment and control populations, using the same seeding density used in step 5.3.5.
- Separately pool the number of cells required for library coverage for each replicate in the drug treatment group. Add the drug at intermediate concentrations (IC20-IC50). Seed the cells and incubate (37 °C, 5% CO2) until next passage.
- Separately pool the number of cells required for library coverage for each replicate in the vehicle control group. Add the vehicle control using the same volume as the drug (<0.5% v/v). Seed the cells and incubate (37 °C, 5% CO2) until the next passage.
- Continue to passage the cells and harvest the cell pellets for genomic DNA every 3 days as described in step 5.3.5, while refreshing the drug or vehicle at each passage.
- For the positive selection or drug resistance screens, split each replicate group according to the number of cells required for library coverage. Add IC90 drug concentrations to each replicate. At IC90, a majority of cells will be killed. Allow resistant populations to grow and collect cell pellets (1–2 x 107 cells) for genomic DNA extraction.
6. CRISPR sample preparation and sequencing
- Genomic DNA purification
- Incubate the frozen cell pellets for 5–10 min at RT for thawing.
- Add 1.4 mL of PBS to a 50 mL centrifuge tube containing a cell pellet. Vortex for 20 s to resuspend the cells and rest for 1 min. If required, pipette 15x with P1000 to break up the remaining cell clumps. If transferring cells from a 15 mL or 1.5 mL tube, resuspend the cells with 1 mL of PBS, then transfer cells to a 50 mL tube and rinse the original tube with 400 µL of PBS.
- Add 5 mL of Nuclei Lysis Solution to the resuspended cells. Using a 10 mL pipette, mix the sample by pipetting up and down 5x.
- Add 32 µL of RNase A (20 mg/mL; to obtain a final concentration of 100 µg/mL) to the nuclear lysate and mix the sample by inverting the tube 5x. Incubate the mixture at 37°C for 15 min and allow sample to cool for 10 min at RT.
- Add 1.67 mL of Protein Precipitation Solution to the lysate and vortex vigorously for 20 s. Small protein clumps may be visible after mixing.
- Centrifuge at 4,500 x g for 10 min at RT.
- Using a 10 mL pipette, transfer the supernatant to a 50 mL centrifuge tube containing 5 mL of isopropanol. Gently mix the solution 10x by inversion until the DNA is observed.
NOTE: DNA can be observed as white, thread-like strands that form a visible mass. - Centrifuge at 4,500 x g for 5 min at RT to pellet the DNA.
- Using a 10 mL pipette, carefully remove the supernatant and avoid dislodging the DNA pellet. Add 5 mL of 70% ethanol at RT to the DNA. Gently rotate the tube to wash the DNA pellet and sides of the centrifuge tube.
- Centrifuge at 4,500 x g for 5 min at RT.
- Using a 10 mL pipette, carefully remove the 70% ethanol and avoid dislodging the DNA pellet. Air-dry genomic DNA for 10 min at RT.
- Add 400 µL of TE solution to the tube and let the DNA dissolve by incubating at 65 °C for 1 h. Mix the DNA by gently flicking the tube every 15 min. If the DNA does not dissolve completely, incubate tube at 65 °C for an additional 1 h while gently flicking the tube every 15 min, and leave it at 4 °C overnight.
- Centrifuge at 4,500 x g for 1 min at RT and transfer genomic DNA to a 1.5 mL low-binding tube.
- Quantify and measure the purity of genomic DNA on both the spectrophotometer (for total nucleic acid content) and fluorometer (for double-stranded DNA content).
- Optionally, precipitate genomic DNA if there are issues with downstream PCR amplification of the sgRNA as follows.
- Transfer 400 µL genomic DNA into a 1.5 mL microcentrifuge tube.
- Add 18 µL of 5 M NaCl (final concentration of 0.2 M) and 900 µL of 95% ethanol.
- Invert tube 10x until thoroughly mixed, then centrifuge at 16,000 x g for 10 min at RT.
- Carefully remove the supernatant and avoid dislodging the DNA pellet. Wash the DNA pellet with 500 µL of 70% ethanol. Gently rotate the tube to wash the DNA pellet.
- Centrifuge at 16,000 x g for 5 min at RT.
- Carefully remove supernatant and avoid dislodging DNA pellet. Air-dry genomic DNA for 10 min at RT.
- Add 300 µL of TE to dissolve DNA as described in steps 6.1.12.
- Quantify and measure the purity of genomic DNA as described in step 6.1.14.
- CRISPR sequencing library preparation
- Set up PCR 1 as outlined in Table 5 using a total of 100 μg of genomic DNA. Add 3.5 μg of genomic DNA per 50 μL reaction and set up identical 50 μL reactions to achieve the desired coverage. Table 6 lists examples of primer sequences for amplification of LCV2::TKOv3 sequencing libraries. Table 7 lists examples of primer sequences for amplification of pLCKO2::TKOv3 sequencing libraries.
- Amplify PCR 1 reactions in a thermocycler using the program outlined in Table 8.
- Check PCR 1 amplification by running 2 μL of the PCR product on a 1% agarose gel. PCR 1 yields a product of 600 bp.
- Pool all individual 50 μL reactions for each genomic DNA sample and mix by vortexing.
- Set up one PCR 2 reaction (50 μL) for each sample as outlined in Table 9 using 5 μL of the pooled PCR 1 product as template. Use unique index primer combinations for each individual sample to allow pooling of sequencing library samples.
- Amplify the PCR 2 reaction in a thermocycler using the program outlined in Table 10.
- Clean agarose gel equipment for purifying amplified products with 0.1 N HCl for 10 min prior to casting a gel. Prepare a 2% agarose gel containing DNA stain for purifying PCR 2 amplified products.
- Run the PCR 2 product on the 2% agarose gel at low voltage (1.0–1.5 h run). PCR 2 yields a product of 200 bp.
- Visualize the PCR products on a blue light transilluminator. Excise the 200 bp band and purify DNA from the agarose gel slice using a gel extraction kit. Quantify and measure the purity of the sequencing library on both the spectrophotometer and fluorometer.
NOTE: A typical gel-purified sequencing library concentration ranges from 5–10 ng/μL and a total yield of 150–300 ng.
- High-throughput sequencing
- Sequence the CRISPR sequencing libraries on next-generation sequencers.
- Sequence reference T0 samples at higher read depth of 400- to 500-fold library coverage. Sequence experimental timepoint samples for drop-out screens at a minimum read depth of 200-fold. For strong positive selection screens, a minimum of read depth of 50-fold coverage is sufficient for identification of enriched sgRNAs.
NOTE: It is critical to sequence the T0 sample to determine library representation for a particular screen and serve as a reference for the determining sgRNA fold changes over time.
7. Data analysis
- Align sequence using programs such as Bowtie to map sequence reads to the reference library using the following parameters: -v2 (allowing two mismatches) and -m1 (discarding any read that mapped to more than one sequence in the library).
- Normalize the number of uniquely mapped reads for each sgRNA for a given sample to 10 million reads per sample as follows:
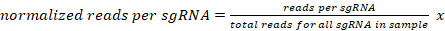 107
107 - Calculate the log2 fold change of each sgRNA for each replicate at each timepoint (Tn) compared to the T0 sample (Tn/T0). Add a pseudo count of 0.5 reads to all read counts to prevent discontinuities from zeros. Exclude sgRNAs with <30 raw reads in the T0 sample from fold-change calculation and downstream analysis.
- Analyze fold changes with the Bayesian Analysis of Gene Essentiality (BAGEL) algorithm <https://github.com/hart-lab/bagel>, using the core essential and non-essential training sets defined previously19 for gene essentiality screens (Supplementary Table S1) or DrugZ <https://github.com/hart-lab/drugZ> for drug screens20.
- Calculate the precision and recall for screen performance assessment using BF scores. Use the essential set from step 7.4 as the true positive list for the precision_recall_curve function of the Scikit-learn library for Python, along with the above BF score subset. Alternatively, perform the same using the PRROC package in R.
- Calculate the mean fold change of all guides for each gene. Generate density plots for the essential and non-essential genes (see step 7.4) in R or equivalent software. In R, if x.ess is a vector containing the log fold change values of essential genes and x.nonEss contain non-essential genes, plot using the following command:
plot( density( x.ess ), xlab=”mean logFC”,col=”red”,lwd=2 )
lines( density( x.nonEss ), col=”blue”,lwd=2 )
NOTE: For Python version details and packages used, see scikit-learn v0.19.1: (published by Pedregosa et al.21).
Representative Results
Overview of genome-scale CRISPR screening workflow
Figure 1 illustrates an overview of the pooled CRISPR screening work flow, starting with infection of target cells with CRISPR library lentivirus at a low MOI to ensure single integration events and adequate library representation (typically 200- to 1000-fold). Following infection, cells are treated with the antibiotic puromycin to select for transduced cells. After selection, a baseline T0 cell pellet is collected to assess library distribution at the start of screening. The remaining cells, comprised of a heterogeneous population of genetic perturbations, are passaged at desired library representation every 3-4 days for 15-20 doublings to allow gene editing and the resulting effects to manifest. Screens with drug treatments are typically added at T3 or T6 after the cells have recovered from virus infection and puromycin selection. Cells are harvested at the desired library representation at every passage for genomic DNA, to determine guide abundance by next generation sequencing at desired timepoints.
It is recommended to collect multiple samples in case of any failures that may occur in the downstream sequencing library preparation steps. Pooled screens are typically viability-based assays that are designed for either positive or negative selection of essential sgRNAs. Positive screens identify genes that show resistance or increase survival under specific selection pressure (e.g., drugs or mutant cell line). In this case, most cells will die from the selection, and cells that remain will be enriched for sgRNAs targeting genes that are resistant for the drug or condition being tested. Negative selection screens or “drop-out” screens identify gene knock-outs with increased sensitivity to or loss of survival under the screen selection pressure. To identify perturbations that have a phenotypic effect such as a growth defect, guide abundance at each timepoint is quantified by next-generation sequencing and compared to T0 to assess drop-out or enrichment of guides over the course of the screen. Using analysis platforms, log-fold changes are measured for guides, and algorithms such as the BAGEL can be applied to enable ranking of gene hits.
Library amplification and maintenance of library representation in pooled CRISPR screens
Figure 2 illustrates the expected distribution of guides after amplification of the plasmid library. TKOv3 library consists of 71,090 sgRNAs with four sgRNAs per gene, targeting ~18,000 protein coding genes10. An ideal library should have every single sgRNA represented at similar quantities. Therefore, it is recommended to confirm the distribution of guides in the amplified library by next-generation sequencing. Shown here is an amplified library with very tight distribution of sgRNAs, confirming that >95% of all sgRNAs are within 4-fold distribution range (Figure 2). A wider distribution of sgRNAs will indicate that the abundance of library guides are not equally represented and can contribute to the noise in pooled screens.
Evaluation of screen performance
Figure 3 illustrates that the performance quality of a screen can be evaluated by assessing the fold change distribution of all sgRNA against a gold standard reference list of essential (684 genes) and nonessential genes (926 genes) and visualized as precision-recall curves10. Using the gold-standard reference sets, Bayes Factor (BF) scores are calculated for the screen endpoint, and precision-recall curves are plotted. BF scores are calculated by analyzing the log-fold change for all guides targeting a gene using a Bayesian framework (the BAGEL algorithm described previously19) to compare distributions of known essential and non-essential guide sets. False discovery rates (FDR) are derived empirically using the same gold standard reference sets. A high performing screen should recover a high number of essential genes at a threshold of BF >6 and FDR <5%, as evidenced by a sharp “elbow” in most curves and a straight line to the terminal point as shown by the blue line in Figure 3A. The dropout of guides targeting essential and nonessential genes should also be examined (Figure 3B). Guides targeting the reference nonessential genes should show a largely symmetric distribution of log-fold changes centered at zero, as shown by the dashed line in Figure 3B. The fold change distribution of guides targeting essential genes shows a strong negative shift relative to the distribution of guides targeting nonessential genes, as shown by the solid line in Figure 3B.
Essential genes
One of the basic applications of pooled genome-wide drop-out screens is to identify essential genes. Essential genes, a subcategory of fitness genes, are genes whose perturbation causes cell lethality, also considered loosely as proliferation genes. In the context of cancer biology, it is possible to identify context-specific essentials in order to identify dependencies for a particular tumor cell line. Figure 4, shows the gene rank of essential genes using Bayes Factor scores, derived from the BAGEL algorithm. Bayes Factor (BF) represents a confidence measure that the gene knock-out results in a fitness defect. More positive scores indicate higher confidence that the perturbation causes a decrease in fitness.
Positive selection screen
Genome-wide knock-out pools can be cultured in the presence of excess drug agent to look for suppressor/resistance genes. Shown here is an example of HCT116 cells screened in the presence of thymidine to look for suppressors of G1/S arrest3. Details of this screen can be found in a previous publication3. Briefly, 6 days after selection of CRISPR library infected cells, cells were split into replicates maintaining library coverage and treated with thymidine. Cells were passaged in the presence of drug until ample resistant cells were recovered for genomic DNA sampling. Positive selections can be sequenced (read depth) at lower coverage than negative screens since only a small fraction of guides will remain due to the strong selective pressure. In this example, sequencing was obtained with a few million reads, and 11 of 12 sgRNAs targeting thymidine kinase (TK1) were recovered and enriched as expected (Figure 5).
| Amounts were determined based on molar ratio of 1:1:1 | ||
| Component | Amount per 15-cm plate a | |
| LCV2::TKOv3 | pLCKO2::TKOv3 | |
| psPAX2 | 4.8 µg | 7.0 µg |
| pMD2.G | 3.8 µg | 4.0 µg |
| TKOv3b | 8.0 µg | 5.0 µg |
| aAmounts determined based on most productive plasmid combination for TKO library at 1:1:1 molar ratio | ||
| bAmount TKO plasmid based on CRISPR library vector backbone. LCV2 all-in-one vector =13 kb, non-Cas9 pLCKO2 vector = 7.6 kb | ||
Table 1: Recommended amount of plasmid for TKOv3 transfection.
| Component | Amount per 15-cm plate |
| Opti-MEM | 800 µL |
| Transfection reagent | 48 µL |
Table 2: Lipid-based transfection reagent set-up.
| Fold-coverage | Number of cells per sgRNAb | Number of cells required for infectionb |
| (sgRNA library sizea × fold coverage) | (sgRNA library size × fold coverage ÷ 0.3 MOI) | |
| 200 | 1.5 x 107 | 5 x 107 |
| 500 | 3.6 x 107 | 1.2 x 108 |
| 1000 | 7.1 x 107 | 2.4 x 108 |
| a Based on TKOv3 library size = 71,090 sgRNA | ||
| b Numbers are rounded up | ||
Table 3: Determination of cell numbers required for TKOv3 CRISPR library infection and cell plating at various fold-coverage.
| Treatment | Number of plates required for infection | |
| Screening plates | Virus, + puromycin | (sgRNA library size × 200-fold) ÷ 0.3 MOI ÷ cell seeding density at infection = number of plates requireda |
| Control 1 | No virus, + puromycin (0% survival control) | 1 |
| Control 2 | Virus, + No puromycin (100% survival control) | 1 |
| a Include extra plates to accommodate for MOI fluctuations and growth rates | ||
Table 4: Calculation for infection set-up.
| Reagents | Amount per 1x reaction |
| 2x Master Mix | 25 μL |
| 10 mM PCR 1 LCV2 forward primer | 2.5 μL |
| 10 mM PCR 1 LCV2 reverse primer | 2.5 μL |
| Genomic DNA | 3.5 μg |
| Water | up to 50 μL |
| Total | 50 μL |
Table 5: PCR 1 set-up.
Table 6: PCR primers for amplification of LCV2::TKOv3 sequencing libraries. Please click here to download this file.
Table 7: PCR primers for amplification of pLCKO2::TKOv3 sequencing libraries. Please click here to download this file.
| Step | Temperature | Time | |
| 1 | 98°C | 30 sec | |
| 2 | 98°C | 10 sec | 25 cycles (step 2 – 4) |
| 3 | 66°C | 30 sec | |
| 4 | 72°C | 15 sec | |
| 5 | 72°C | 2 min | |
| 6 | 10°C | Hold |
Table 8: PCR 1 cycle parameters.
| Reagents | Amount per 1x reaction |
| 2x Master Mix | 25 μL |
| 10 mM i5 forward primer | 2.5 μL |
| 10 mM i7 reverse primer | 2.5 μL |
| PCR 1 product | 5 μL |
| Water | 15 μL |
| Total | 50 μL |
Table 9: PCR 2 set-up.
| Step | Temperature | Time | |
| 1 | 98°C | 30 sec | |
| 2 | 98°C | 10 sec | 10 cycles (step 2 – 4) |
| 3 | 55°C | 30 sec | |
| 4 | 65°C | 15 sec | |
| 5 | 65°C | 5 min | |
| 6 | 10°C | Hold |
Table 10: PCR 2 cycle parameters.
Supplementary table S1. TKO reference gene sets Please click here to download this file.
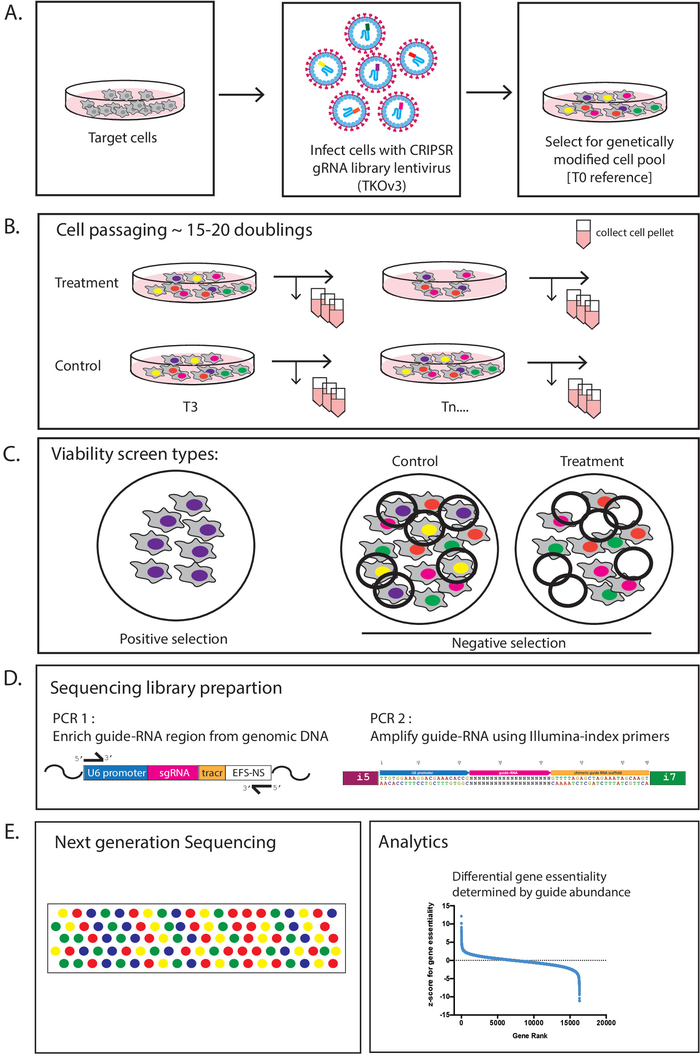
Figure 1: Schematic overview of pooled screening workflow. (A) Target cell population is infected with CRISPR library lentivirus at low MOI to ensure that most cells receive one viral integration and that library representation is maintained. The different colors represent different sgRNAs in each viral particle. Genetically modified cell pools are selected. Once selection is complete, cells are sampled for T0 reference and serially passaged. (B) At the first passage after T0, cells have recovered from infection and drug treatments can be added, if required. Following treatment, cell populations are serially passaged for several weeks. During each passage, cells are collected for genomic DNA and reseeded at the required fold coverage of the sgRNA library. (C) Two types of screens can be performed: 1) positive selection screens, which identify mutant cells that show resistance or increased survival under the specific selection pressure (e.g., drugs or mutant cell line), as they will be enriched during the screen; or 2) negative selection screens, which identify mutant cells with increased sensitivity to or loss of survival under the screen selection pressure, as they will be lost during the screen. (D) Genomic DNA is harvested and PCR-amplified to enrich for guide regions. (E) Guide abundance is quantified by next-generation sequencing and enriched, or depleted guides are determined for “hit” identification. Please click here to view a larger version of this figure.
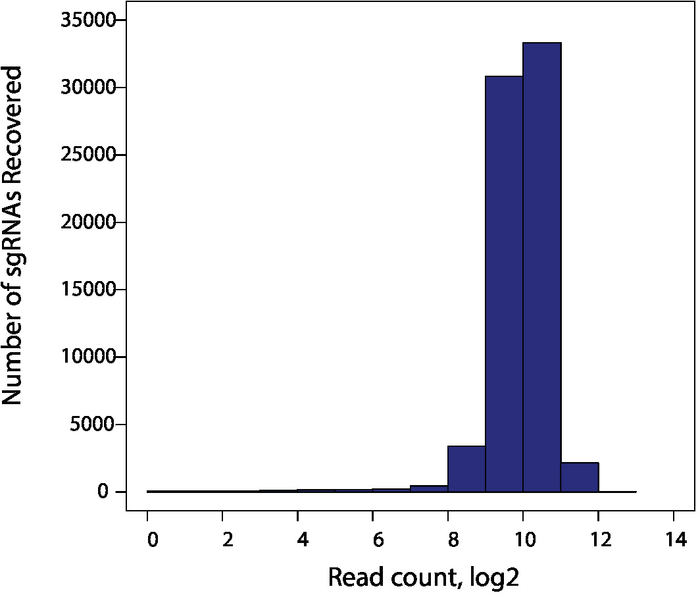
Figure 2: Quality of amplified CRISPR sgRNA library. Amplified library plasmids are analyzed by next-generation sequencing (recommended reads: 30 million reads, corresponding to ~400-fold representation of the library). Shown here is a library with tight distribution of sgRNAs, with >95% of all sgRNAs within a 4-fold distribution range. Please click here to view a larger version of this figure.
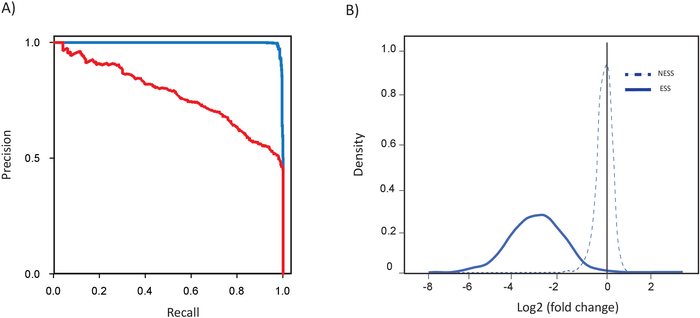
Figure 3: Evaluation of drop-out screen quality using gold-standard essential gene reference sets. (A) Precision recall analysis of screening results in recovering of essential genes at a threshold of BF >6 and FDR of 5%. High performing screen are represented by blue line and low performing screens are represented by red line. (B) Fold change distribution of sgRNA targeting essential genes (solid line) and nonessential genes (dotted lines). Please click here to view a larger version of this figure.
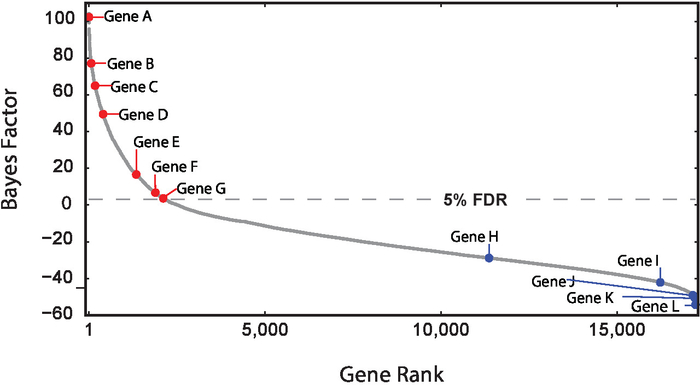
Figure 4: Determination of gene essentiality. Bayes Factor ranking of gene essentially in a particular screen. Bayes Factor (BF) represents a confidence measure that the gene knock-out results in a fitness defect. Higher Bayes Factors indicate increased confidence that gene knock-out results in fitness defect, (red dots). Lower Bayes Factor scores suggest knock-out provides growth advantage (blue dots). Please click here to view a larger version of this figure.
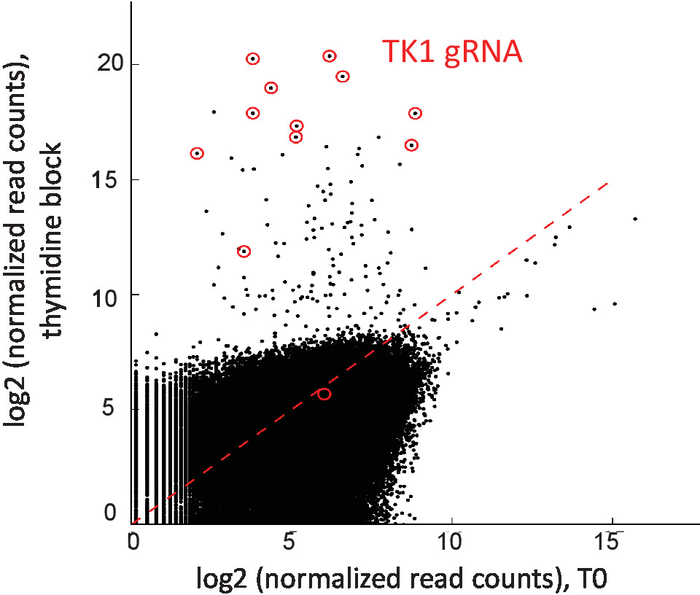
Figure 5: Positive selection screen for suppressor of thymidine block in HCT116 cells. Normalized read counts for all sgRNAs at T0 plotted against mean normalized read counts for thymidine treated samples. For positive selection screens (i.e., using an IC90 concentration of drug), the number of perturbations that will confer resistance to the drug is expected to be small. For this reason, read depth can be lower than what is needed for negative screens, in whch most of the library is expected to be represented. TK1 sgRNAs are circled in red. This figure has been modified from a previous publication3. Please click here to view a larger version of this figure.
Discussion
Due to its simplicity of use and high pliability, CRISPR technology has been widely adopted as the tool of choice for precise genome editing. Pooled CRISPR screening provides a method to interrogate thousands of genetic perturbations in a single experiment. In pooled screens, sgRNA libraries serve as molecular barcodes, as each sequence is unique and is mapped to the targeted gene. By isolating the genomic DNA from the cell population, genes causing the phenotype of interest can be determined by quantifying sgRNA abundance by next generation sequencing. Massively parallel sequencing methods are utilized to quantify sgRNAs in samples, meaning that multiple independent cell populations can be pooled into the same sequencing lane to minimize cost.
Before embarking on a large-scale screening project, it is important to have a well-characterized and technically optimized model. Genetic background, growth rate, and transduction efficiency are important factors when choosing your cell lines for screening. For example, growth rates and editing efficiency will determine scalability and technical suitability of the model. In order to adequately represent large sgRNA libraries, tens of millions of cells are required, therefore cell number could be a limiting factor in screening feasibility for cell lines with slower doublings or ones that do not have good proliferative capacity (e.g., primary cells). Based on growth rates, cell culture conditions such as cell seeding density and plate size for screening should be selected accordingly. It is recommended to culture cells in the largest vessel that is practical and technically feasible for the screen.
Lentivirus transduction efficiencies vary between cell types, as cells differ in inherent infectivity. As a result, the volume of virus required to achieve sufficient infection in one cell type will not necessarily be the same in another. Therefore, it is critical to functionally titer each batch of lentivirus library produced in the cell line to be screened to ensure sufficient coverage of the library and mostly single transduction events per cell by transducing at lower MOIs around 0.3 (section 4). Transduction efficiencies can also be influenced by cell culture conditions; therefore, functional titers should be determined using the same cell conditions that will be used in the screen. That is, it is important to use the same tissue culture vessels, media constituents and volume, cell plating density, and virus preps without prior thaws. Measurements made in different formats or conditions will not reliably scale to the screening format.
Despite the advantage of using all-in-one CRISPR-Cas9 guide libraries such as LCV2::TKOv3, the gene encoding Cas9 is quite large, making it difficult to efficiently package into viral particles (105-106 TU/mL). Delivering lower lentiviral titers can be a limitation for cell lines that are difficult to transduce, as they will have even more difficulty with the all-one-CRISPR libraries. To mitigate this, Cas9 should be expressed in the cell line in advance, followed by delivery of CRISPR libraries only containing sgRNAs (e.g., pLCKO2::TKOv3), which can be made at much higher titers (107-108 TU/mL). The ploidy of a cell line is also important, as it determines the number of target loci that need to be modified. The ability to generate complete knock-outs in haploid cells is more efficient than in cells with multiple copies of a given gene. Therefore, screens in haploid cells may be more sensitive and yield higher quality data than screens performed in diploid or aneuploid cell lines6. Testing known genes that are linked to the phenotype will help determine the screen-ability of a cell line model. For example, for essentiality screens, guides targeting a subunit of the 26S proteasome, PSMD1 (Addgene: plasmid #74180), a core essential gene, can be used to test editing efficiency and infectibility of cell lines, as perturbation of PSMD1 will result in cell death.
The robustness of pooled screens highly depends on sgRNA representation. This is an important metric that determines library performance during a screen and the ability to identify hits. Library diversity is biased in the representation of each sgRNA; therefore, the population of cells to be screened and analyzed should be sufficiently large to ensure the capture of under-represented sgRNAs6. 200- to 1000-fold representation of each sgRNA is the typical coverage that has been used in published screens (i.e., 200-1000 cells per sgRNA)10,15. This representation should be maintained when amplifying the library plasmid (section 1) and throughout the screen by infecting and passaging the required cell number (section 5) to represent the desired library coverage and during sequencing library preparation (protocol 6), as described throughout the protocol. For example, to achieve ~200-fold coverage of the TKOv3 library requires selection and passaging of 15 million infected cells. During sequencing, assuming a diploid human genome contains ~7.2 pg of DNA and 1 sgRNA per genome, a total of 100 μg of genomic DNA is required to generate the sequencing library for 15 million sequence reads. The decision of coverage will depend on the size of the library, as coverage of larger libraries will require culturing larger number of cells that can be difficult to maintain and not technically practical. A minimum of 200-fold coverage is recommend with TKOv3 libraries, as 200-fold provides an optimal balance between the logistics of screening large number of cells and maintaining sufficient dynamic range to detect true biological sgRNA drop-outs with limited noise from random depletions22,23. Higher fold library representations will result in improved reproducibility and ensure sufficient window for detection of changes in sgRNA abundance, especially for negative selections. A limiting feature of negative screens is that the perturbation is only depleted to the extent that it was present in the starting library24. In comparison, the dynamic range of positive selection screens is much larger, as they rely on enrichment of cells, and could enrich to 100% of the final population23. Therefore, for positive selection screens (e.g. drug resistance screens), library coverage and read depth can be reduced to 50- to 100-fold representation since only a small cell population is expected to survive.
The sequencing library protocol described here is a two-step PCR optimized for TKOv3 CRISPR libraries in both vector backbones and sequenced on the Illumina sequencing platform. These sequencing libraries can also be generated using a single PCR protocol, similar to that described in Hart et al.3. For other ready-made libraries, the primers and sequencing protocols provided for those libraries should be consulted. When preparing genomic DNA and PCR samples, it is essential to be considerate of contamination precautions. For example, a dedicated area for genomic DNA purification is highly recommended. It should also be physically distinct from bacterial plasmid preps, which are common contaminants found in genomic DNA samples. PCR reactions should be set up in a dedicated PCR hood, as this will minimize contamination from plasmids and other sequencing libraries. For good practice, a no-template negative control can be included to help monitor for PCR contamination.
Data analysis to translate sequencing reads from screens is a non-trivial task, given the size and diversity of these datasets. Once the sequence reads have been aligned and normalized, several bioinformatic tools are available to assist with evaluating screen performance (Figure 3) and hit identification (Figure 4). BAGEL is described in this protocol as the key tool for data analysis. BAGEL uses a Bayesian framework to compare the distributions of known essential and non-essential gene sets to the log-fold change of all guides targeting a gene. This method is described in detail in Hart et al3. In addition to BAGEL, other algorithms designed to identify both enriched and depleted sgRNAs, such as MAGeCK25 can also be used. For drug screens, it is recommended to use the DrugZ algorithm to identify both synergistic and suppressor chemical genetic interactions. DrugZ was designed to compare the relative abundance of sgRNA in a treated population to the relative abundance of sgRNA in an untreated population at the same timepoint20.
A limitation of CRISPR screens is that Cas9 does not always lead to a knock-out, as there is always a possibility that the indels created are in-frame mutations, leaving the gene function intact13. This results in a mixed population, making the screen “noisy” and interpretation of data challenging. Using multiple independent sgRNAs targeting a gene can build-in redundancy, reducing the effect of sgRNAs with low activity. An additional caveat to CRISPR studies is the effect of the double strand breaks created by Cas9 nuclease, which can lead to cellular lethality independent of the gene being targeted. This anti-proliferative effect increases with target site copy number, leading to false positive identification of genes within highly amplified regions26. Computational methods like CERES have been developed to correct for copy number effects27. These workflows consider the copy number effect to estimate gene dependency levels in knock-out-based essentiality screens. Careful examination of genomic locations of hit genes in amplified regions can help determine false positives that are due to multiplicity of cutting effects13. Primary screens can only identify potential hits. It is important to follow-up with a secondary screen or protocol to validate the hits and distinguish on-target from off-target effects, weeding out false positives and ensuring genes that scored weakly due to ineffective perturbations are not left behind as false negatives23.
This protocol focuses on viability-based screening approaches, in which the condition of study should lead to a proliferation defect or death of cells. For processes that do not lead to a change in cellular viability, the viability-based pooled screening method can be restrictive. An alternative is to perform screens using reporter or marker-based assays and enrichment by fluorescence activated cell sorting (FACS) approaches. In marker-based selection screens, the phenotype is based on mutations that regulate marker gene expression rather than cell health13,23. Arrayed CRISPR formats are also available for one-gene per well screening. Arrayed formats are more amenable to complex or microscopy-based read outs. However, arrayed formats require automated equipment and large amounts of reagents28.
The screening protocol discussed here uses S. pyogenes Cas9 nuclease to create null alleles, which is the most widely used for genetic screens and for which many libraries are available (Addgene: Pooled Libraries). Alternative options to knock-out libraries are also available, which use a catalytically dead dCas9 tethered to chromatin modifier proteins to inhibit (CRISPRi) or activate (CRISPRa) transcription of genes. Similar to RNAi, CRISPRi offers the ability to study phenotypic effects at different gene doses and essential genes that cannot tolerate complete knock-out, while CRISPRa can be used to perform gain-of-function screens. Each of these technologies have their advantages, but in general, the CRISPR knock-out approach is the most developed. It has been proven to perform well with low noise, minimal off-target effects, and experimental consistency, especially in lethality-based essential gene screens, when compared to knock-down approaches using either CRISPRi and shRNAs5. Despite its extensive applicability to date, CRISPR screening technology remains in its early stages. New tools are continuing to be built from the basic components of CRISPR. These include combinatorial gene editing strategies that can target multiple genomic loci, optimization of orthogonal Cas enzymes, and modifications with chromatin functional domains to diversify Cas9 activities. As CRISPR technology continues to grow, its coupling to genetic screening approaches will serve as a powerful platform for functional discovery in genetics.
Declarações
The authors have nothing to disclose.
Acknowledgements
This work was supported by Genome Canada, the Ontario Research Fund, and the Canadian Institutes for Health Research (MOP-142375, PJT-148802).
Materials
| 0.22 micron filter | |||
| 30°C plate incubator | |||
| 37°C shaking incubator | |||
| 37°C, 5% CO2 incubator | |||
| 5 M NaCl | Promega | V4221 | |
| 50X TAE buffer | BioShop | TAE222.4 | |
| 6 N Hydrochloric acid solution | BioShop | HCL666.500 | |
| 95% Ethanol | |||
| Alamar blue | ThermoFisher Scientific | DAL1025 | |
| Blue-light transilluminator | ThermoFisher Scientific | G6600 | |
| Bovine Serum Albumin,Heat Shock Isolation, Fraction V. Min. 98%, Biotechnology grade | Bioshop | ALB001.250 | |
| Dulbecco's Modification of Eagles Medium | Life Technologies | 11995-065 | Cel culture media |
| Electroporation cuvettes | BTX | 45-0134 | |
| Electroporator | BTX | 45-0651 | |
| Endura electrocompetent cells | Lucigen | 90293 | |
| Fetal Bovine Serum | GIBCO | 12483-020 | |
| HEK293T packaging cells | ATCC | CRL-3216 | recommend passage number <15 |
| Hexadimethrine Bromide (Polybrene) | Sigma | H9268 | Cationic polymer to enhance transduction efficiency |
| Hexadimethrine Bromide (Polybrene) | |||
| LB agar plates with carbenicillin | |||
| LB medium with carbenicillin | |||
| Low molecular weight DNA ladder | New England Biolabs | N3233S | |
| Nanodrop spectrophotometer | ThermoFisher Scientific | ND-ONE-W | |
| NEBNext Ultra II Q5 Master Mix | New England Biolabs | M0544L | |
| Opti-MEM | Life Technologies | 31985-070 | Reduced serum media |
| Plasmid maxi purification kit | Qiagen | 12963 | |
| pMD2.G (envelope plasmid) | Addgene | Plasmid #12259 | lentiviral system |
| psPAX2 (packaging plasmid) | Addgene | Plasmid #12260 | lentiviral system |
| Puromycin | Wisent | 400-160-UG | |
| QIAquick gel extraction kit | Qiagen | 28704 | |
| Qubit dsDNA BR assay | ThermoFisher Scientific | Q32853 | |
| Qubit fluorometer | ThermoFisher Scientific | Q33226 | |
| RNAse A | Invitrogen | 12091021 | |
| S.O.C recovery medium | Invitrogen | 15544034 | |
| SYRB Safe DNA gel stain | ThermoFisher Scientific | S33102 | |
| Toronto KnockOut CRIPSR library (TKOv3) – Cas9 included | Addgene | Addgene ID #90203 | Genome-wide CRISPR library , includes Cas9, 71,090 sgRNA |
| Toronto KnockOut CRIPSR library (TKOv3) – non-cas9 | Addgene | Addgene ID #125517 | Genome-wide CRISPR library, non-Cas9, 71,090 sgRNA |
| Tris-EDTA (TE) solution, pH8.0 | |||
| UltraPure agarose | ThermoFisher Scientific | 16500500 | |
| Wizard genomic DNA purification kit | Promega | A1120 | |
| X-tremeGENE 9 DNA transfection reagent | Roche | 06 365 809 001 | Lipid based transfection reagent |
Referências
- Jiang, F., Doudna, J. A. CRISPR-Cas9 Structures and Mechanisms. Annual Review of Biophysics. 46, 505-529 (2017).
- Baliou, S., et al. CRISPR therapeutic tools for complex genetic disorders and cancer (Review). International Journal of Oncology. 53 (2), 443-468 (2018).
- Hart, T., et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell. 163 (6), 1515-1526 (2015).
- Morgens, D. W., Deans, R. M., Li, A., Bassik, M. C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nature Biotechnology. 34 (6), 634-636 (2016).
- Evers, B., et al. CRISPR knock-out screening outperforms shRNA and CRISPRi in identifying essential genes. Nature Biotechnology. 34 (6), 631-633 (2016).
- Miles, L. A., Garippa, R. J., Poirier, J. T. Design, execution, and analysis of pooled in vitro CRISPR/Cas9 screens. The FEBS Journal. 283 (17), 3170-3180 (2016).
- Wang, T., Wei, J. J., Sabatini, D. M., Lander, E. S. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 343 (6166), 80-84 (2014).
- Wang, T., et al. Identification and characterization of essential genes in the human genome. Science. 350 (6264), 1096-1101 (2015).
- Sanson, K. R., et al. Optimized libraries for CRISPR-Cas9 genetic screens with multiple modalities. Nature Communications. 9 (1), 5416 (2018).
- Hart, T., et al. Evaluation and Design of Genome-Wide CRISPR/SpCas9 Knock-out Screens. G3: Genes|Genomes|Genetics. 7 (8), 2719-2727 (2017).
- Mair, B., Tomic, J., et al. Essential gene profiles for human pluripotent stem cells identify uncharacterized genes and substrate dependencies. Cell Reports. 27 (2), 599-615 (2019).
- Shalem, O., et al. Genome-scale CRISPR-Cas9 knock-out screening in human cells. Science. 343 (6166), 84-87 (2014).
- Sharma, S., Petsalaki, E. Application of CRISPR-Cas9 Based Genome-Wide Screening Approaches to Study Cellular Signalling Mechanisms. International Journal of Molecular Sciences. 19 (4), (2018).
- Steinhart, Z., et al. Genome-wide CRISPR screens reveal a Wnt-FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nature Medicine. 23 (1), 60-68 (2017).
- Wang, T., et al. Gene Essentiality Profiling Reveals Gene Networks and Synthetic Lethal Interactions with Oncogenic Ras. Cell. 168 (5), 890-903 (2017).
- Zimmermann, M., et al. CRISPR screens identify genomic ribonucleotides as a source of PARP-trapping lesions. Nature. 559 (7713), 285-289 (2018).
- Deans, R. M., et al. Parallel shRNA and CRISPR-Cas9 screens enable antiviral drug target identification. Nature Chemical Biology. 12 (5), 361-366 (2016).
- Trinh, T. J. J., Bloom, F., Hirsch, V. STBL2: an Escherichia coli strain for the stable propagation of retroviral clones and direct repeat sequences. Focus. 16, 78-80 (1994).
- Hart, T., Moffat, J. BAGEL: a computational framework for identifying essential genes from pooled library screens. BMC Bioinformatics. 17, 164 (2016).
- Wang, G. Z. M., et al. Identifying drug-gene interactions from CRISPR knock-out screens with drugZ. bioRxiv. , (2017).
- Pedregosa, F. V., G, , et al. Scikit-learn: Machine Learning in Python. Journal of Machine Learning Research. 12, 2825-2830 (2011).
- Ketela, T., et al. A comprehensive platform for highly multiplexed mammalian functional genetic screens. BMC Genomics. 12, 213 (2011).
- Doench, J. G. Am I ready for CRISPR? A user’s guide to genetic screens. Nature Review Genetics. 19 (2), 67-80 (2018).
- Hartenian, E., Doench, J. G. Genetic screens and functional genomics using CRISPR/Cas9 technology. FEBS Journal. 282 (8), 1383-1393 (2015).
- Li, W., et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knock-out screens. Genome Biology. 15 (12), 554 (2014).
- Sheel, A., Xue, W. Genomic Amplifications Cause False Positives in CRISPR Screens. Cancer Discovery. 6 (8), 824-826 (2016).
- Meyers, R. M., et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nature Genetics. 49 (12), 1779-1784 (2017).
- Henser-Brownhill, T., Monserrat, J., Scaffidi, P. Generation of an arrayed CRISPR-Cas9 library targeting epigenetic regulators: from high-content screens to in vivo assays. Epigenetics. 12 (12), 1065-1075 (2017).

